| Full name: bile acid-CoA:amino acid N-acyltransferase | Alias Symbol: BAT | ||
| Type: protein-coding gene | Cytoband: 9q31.1 | ||
| Entrez ID: 570 | HGNC ID: HGNC:932 | Ensembl Gene: ENSG00000136881 | OMIM ID: 602938 |
| Drug and gene relationship at DGIdb | |||
Expression of BAAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BAAT | 570 | 206913_at | -0.0214 | 0.9502 | |
| GSE20347 | BAAT | 570 | 206913_at | 0.0528 | 0.4781 | |
| GSE23400 | BAAT | 570 | 206913_at | -0.0738 | 0.0246 | |
| GSE26886 | BAAT | 570 | 206913_at | 0.1519 | 0.2955 | |
| GSE29001 | BAAT | 570 | 206913_at | -0.1509 | 0.2326 | |
| GSE38129 | BAAT | 570 | 206913_at | -0.0389 | 0.6211 | |
| GSE45670 | BAAT | 570 | 206913_at | 0.1907 | 0.0739 | |
| GSE53622 | BAAT | 570 | 124745 | 0.0769 | 0.6960 | |
| GSE53624 | BAAT | 570 | 124745 | 0.3456 | 0.0337 | |
| GSE63941 | BAAT | 570 | 206913_at | 0.1299 | 0.4244 | |
| GSE77861 | BAAT | 570 | 206913_at | -0.0700 | 0.4675 | |
| SRP133303 | BAAT | 570 | RNAseq | 0.0468 | 0.7862 | |
| SRP159526 | BAAT | 570 | RNAseq | 1.7651 | 0.0079 | |
| SRP193095 | BAAT | 570 | RNAseq | 0.1428 | 0.2979 | |
| SRP219564 | BAAT | 570 | RNAseq | -0.0989 | 0.8444 | |
| TCGA | BAAT | 570 | RNAseq | 1.3771 | 0.0260 |
Upregulated datasets: 2; Downregulated datasets: 0.
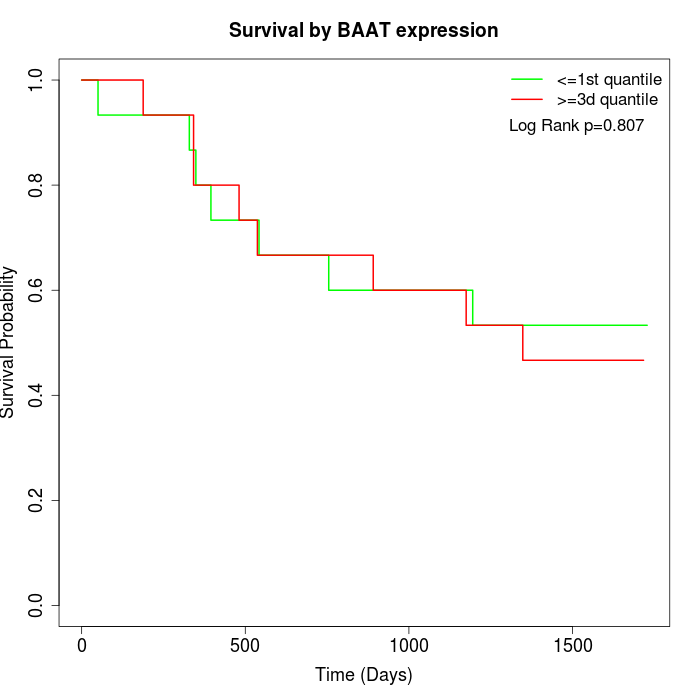
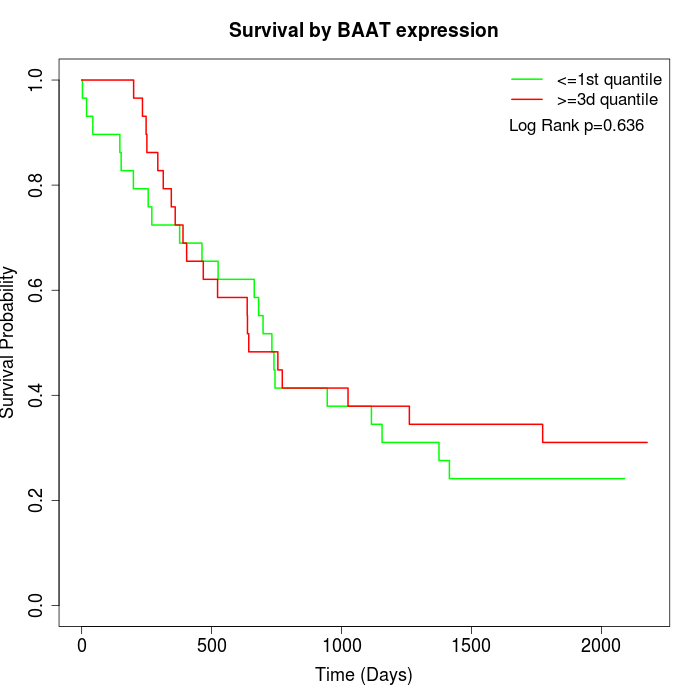
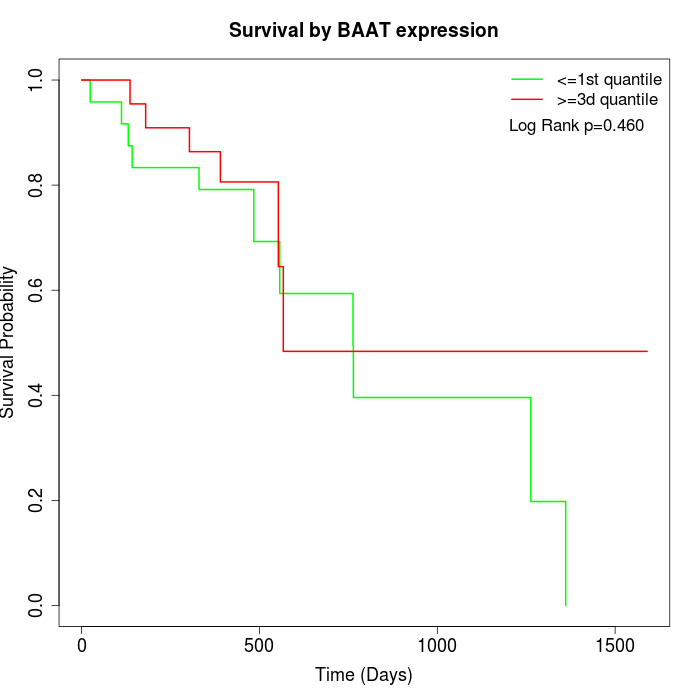
Survival by BAAT expression:
Note: Click image to view full size file.
Copy number change of BAAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BAAT | 570 | 4 | 10 | 16 | |
| GSE20123 | BAAT | 570 | 5 | 9 | 16 | |
| GSE43470 | BAAT | 570 | 6 | 3 | 34 | |
| GSE46452 | BAAT | 570 | 6 | 13 | 40 | |
| GSE47630 | BAAT | 570 | 1 | 17 | 22 | |
| GSE54993 | BAAT | 570 | 3 | 3 | 64 | |
| GSE54994 | BAAT | 570 | 9 | 11 | 33 | |
| GSE60625 | BAAT | 570 | 0 | 0 | 11 | |
| GSE74703 | BAAT | 570 | 5 | 3 | 28 | |
| GSE74704 | BAAT | 570 | 2 | 6 | 12 | |
| TCGA | BAAT | 570 | 28 | 21 | 47 |
Total number of gains: 69; Total number of losses: 96; Total Number of normals: 323.
Somatic mutations of BAAT:
Generating mutation plots.
Highly correlated genes for BAAT:
Showing top 20/187 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BAAT | NFAM1 | 0.685217 | 3 | 0 | 3 |
| BAAT | SYT5 | 0.652056 | 3 | 0 | 3 |
| BAAT | CRB1 | 0.650414 | 3 | 0 | 3 |
| BAAT | PDE6G | 0.632154 | 4 | 0 | 3 |
| BAAT | RITA1 | 0.629252 | 3 | 0 | 3 |
| BAAT | PPP2R5B | 0.624125 | 3 | 0 | 3 |
| BAAT | CHRNA4 | 0.617008 | 3 | 0 | 3 |
| BAAT | ZNF547 | 0.614284 | 3 | 0 | 3 |
| BAAT | CCDC106 | 0.612332 | 3 | 0 | 3 |
| BAAT | DCC | 0.602399 | 3 | 0 | 3 |
| BAAT | HNF4G | 0.594498 | 3 | 0 | 3 |
| BAAT | CRYGD | 0.587223 | 6 | 0 | 5 |
| BAAT | SPX | 0.587118 | 4 | 0 | 3 |
| BAAT | RBMXL2 | 0.586526 | 3 | 0 | 3 |
| BAAT | ELAVL3 | 0.586264 | 5 | 0 | 3 |
| BAAT | FNDC8 | 0.586226 | 5 | 0 | 3 |
| BAAT | KRT76 | 0.585333 | 4 | 0 | 3 |
| BAAT | NIPSNAP1 | 0.584613 | 3 | 0 | 3 |
| BAAT | SLC10A1 | 0.580175 | 5 | 0 | 4 |
| BAAT | C11orf16 | 0.579251 | 7 | 0 | 4 |
For details and further investigation, click here