| Full name: BEN domain containing 4 | Alias Symbol: FLJ35632|FLJ43965 | ||
| Type: protein-coding gene | Cytoband: 4p13 | ||
| Entrez ID: 389206 | HGNC ID: HGNC:23815 | Ensembl Gene: ENSG00000188848 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of BEND4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BEND4 | 389206 | 238101_at | -0.0415 | 0.8865 | |
| GSE26886 | BEND4 | 389206 | 238101_at | 0.1100 | 0.2901 | |
| GSE45670 | BEND4 | 389206 | 238101_at | 0.0873 | 0.3134 | |
| GSE53622 | BEND4 | 389206 | 32900 | -0.2129 | 0.1609 | |
| GSE53624 | BEND4 | 389206 | 105276 | -0.3333 | 0.0084 | |
| GSE63941 | BEND4 | 389206 | 238101_at | 0.0880 | 0.5262 | |
| GSE77861 | BEND4 | 389206 | 238101_at | -0.1686 | 0.1407 | |
| GSE97050 | BEND4 | 389206 | A_24_P914625 | 0.3695 | 0.2237 | |
| SRP133303 | BEND4 | 389206 | RNAseq | -1.2846 | 0.0064 | |
| TCGA | BEND4 | 389206 | RNAseq | -0.7469 | 0.2353 |
Upregulated datasets: 0; Downregulated datasets: 1.
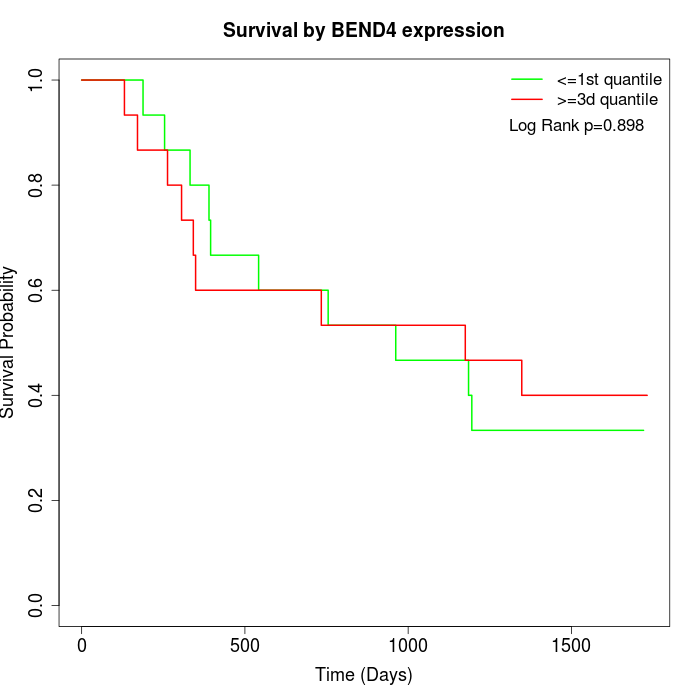
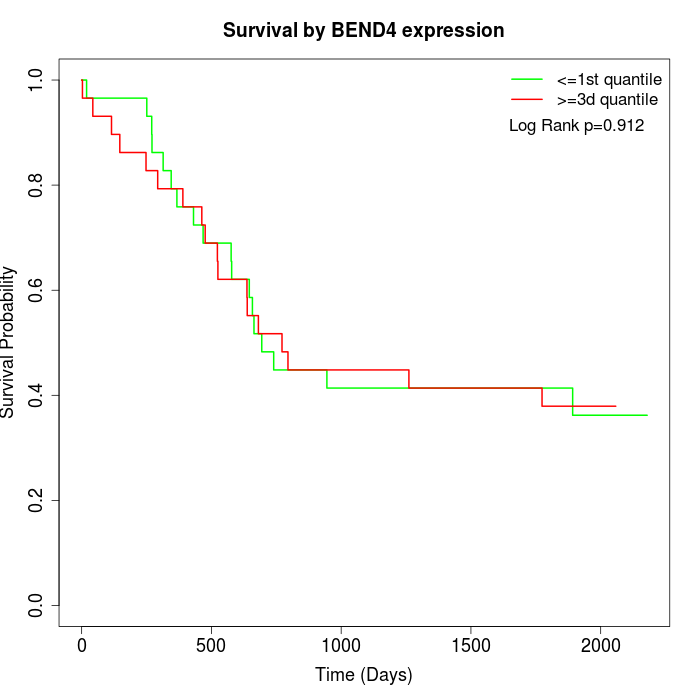
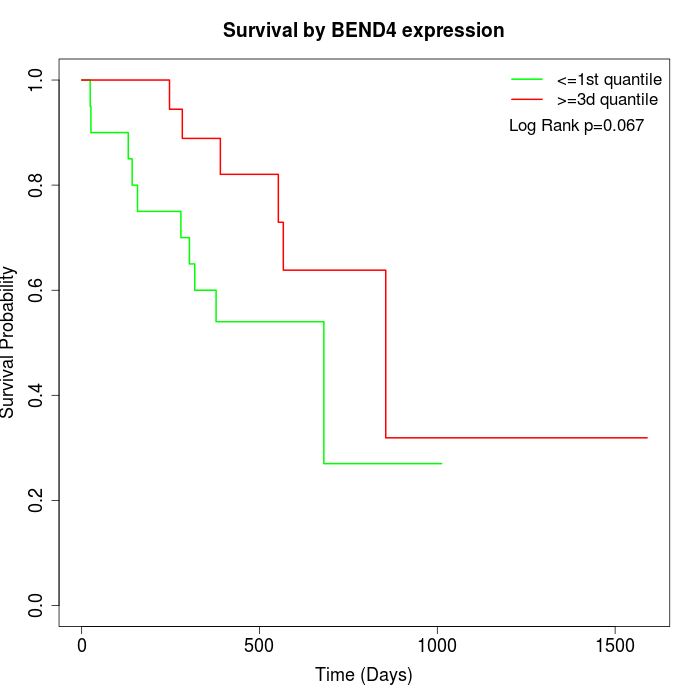
Survival by BEND4 expression:
Note: Click image to view full size file.
Copy number change of BEND4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BEND4 | 389206 | 0 | 16 | 14 | |
| GSE20123 | BEND4 | 389206 | 0 | 16 | 14 | |
| GSE43470 | BEND4 | 389206 | 0 | 14 | 29 | |
| GSE46452 | BEND4 | 389206 | 1 | 36 | 22 | |
| GSE47630 | BEND4 | 389206 | 1 | 18 | 21 | |
| GSE54993 | BEND4 | 389206 | 7 | 0 | 63 | |
| GSE54994 | BEND4 | 389206 | 4 | 9 | 40 | |
| GSE60625 | BEND4 | 389206 | 0 | 0 | 11 | |
| GSE74703 | BEND4 | 389206 | 0 | 12 | 24 | |
| GSE74704 | BEND4 | 389206 | 0 | 9 | 11 | |
| TCGA | BEND4 | 389206 | 11 | 42 | 43 |
Total number of gains: 24; Total number of losses: 172; Total Number of normals: 292.
Somatic mutations of BEND4:
Generating mutation plots.
Highly correlated genes for BEND4:
Showing top 20/177 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BEND4 | IQSEC3 | 0.693238 | 3 | 0 | 3 |
| BEND4 | SSMEM1 | 0.691803 | 3 | 0 | 3 |
| BEND4 | DEFA6 | 0.690078 | 3 | 0 | 3 |
| BEND4 | P2RY4 | 0.687514 | 3 | 0 | 3 |
| BEND4 | SWI5 | 0.680722 | 3 | 0 | 3 |
| BEND4 | KIR2DS3 | 0.678753 | 3 | 0 | 3 |
| BEND4 | TNP1 | 0.678204 | 3 | 0 | 3 |
| BEND4 | CRB2 | 0.676844 | 3 | 0 | 3 |
| BEND4 | KRT20 | 0.672595 | 3 | 0 | 3 |
| BEND4 | NKAIN4 | 0.668744 | 3 | 0 | 3 |
| BEND4 | MC3R | 0.667975 | 3 | 0 | 3 |
| BEND4 | KRT9 | 0.666116 | 3 | 0 | 3 |
| BEND4 | OVCH1-AS1 | 0.664664 | 3 | 0 | 3 |
| BEND4 | SNAI1 | 0.663367 | 3 | 0 | 3 |
| BEND4 | DCD | 0.661758 | 3 | 0 | 3 |
| BEND4 | CABP4 | 0.646209 | 3 | 0 | 3 |
| BEND4 | CACNA1A | 0.645279 | 4 | 0 | 3 |
| BEND4 | RASSF7 | 0.644905 | 3 | 0 | 3 |
| BEND4 | CCDC33 | 0.643876 | 3 | 0 | 3 |
| BEND4 | TM6SF2 | 0.640475 | 4 | 0 | 3 |
For details and further investigation, click here