| Full name: keratin 9 | Alias Symbol: EPPK|K9|CK-9 | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 3857 | HGNC ID: HGNC:6447 | Ensembl Gene: ENSG00000171403 | OMIM ID: 607606 |
| Drug and gene relationship at DGIdb | |||
Expression of KRT9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KRT9 | 3857 | 208188_at | 0.0739 | 0.8988 | |
| GSE20347 | KRT9 | 3857 | 208188_at | 0.2697 | 0.0422 | |
| GSE23400 | KRT9 | 3857 | 208188_at | -0.0436 | 0.4086 | |
| GSE26886 | KRT9 | 3857 | 208188_at | 0.4949 | 0.0149 | |
| GSE29001 | KRT9 | 3857 | 208188_at | 0.6030 | 0.1128 | |
| GSE38129 | KRT9 | 3857 | 208188_at | 0.2984 | 0.0520 | |
| GSE45670 | KRT9 | 3857 | 208188_at | 0.6021 | 0.0155 | |
| GSE63941 | KRT9 | 3857 | 208188_at | 0.4486 | 0.0605 | |
| GSE77861 | KRT9 | 3857 | 208188_at | -0.0131 | 0.9391 | |
| GSE97050 | KRT9 | 3857 | A_23_P15734 | 0.4844 | 0.1838 | |
| SRP159526 | KRT9 | 3857 | RNAseq | 1.1295 | 0.0871 | |
| TCGA | KRT9 | 3857 | RNAseq | 4.0893 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
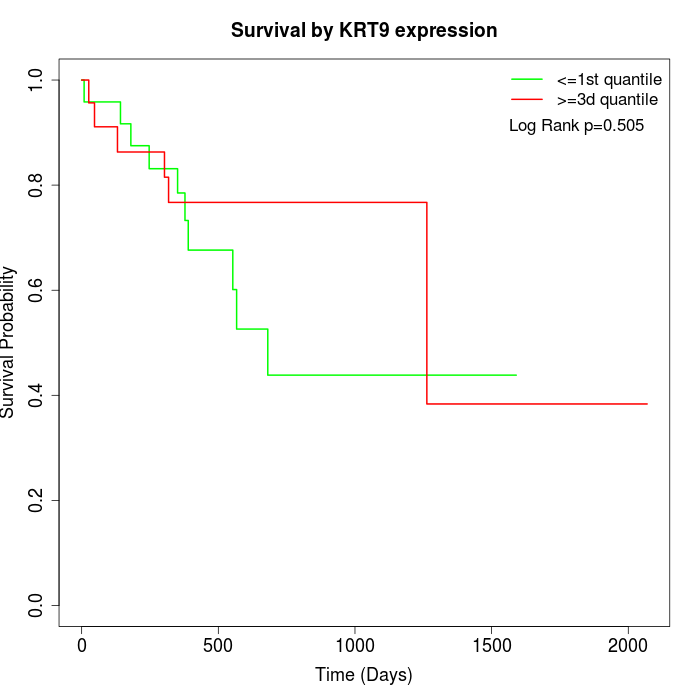
Survival by KRT9 expression:
Note: Click image to view full size file.
Copy number change of KRT9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KRT9 | 3857 | 6 | 2 | 22 | |
| GSE20123 | KRT9 | 3857 | 6 | 2 | 22 | |
| GSE43470 | KRT9 | 3857 | 1 | 2 | 40 | |
| GSE46452 | KRT9 | 3857 | 34 | 0 | 25 | |
| GSE47630 | KRT9 | 3857 | 8 | 1 | 31 | |
| GSE54993 | KRT9 | 3857 | 3 | 4 | 63 | |
| GSE54994 | KRT9 | 3857 | 8 | 5 | 40 | |
| GSE60625 | KRT9 | 3857 | 4 | 0 | 7 | |
| GSE74703 | KRT9 | 3857 | 1 | 1 | 34 | |
| GSE74704 | KRT9 | 3857 | 4 | 1 | 15 | |
| TCGA | KRT9 | 3857 | 23 | 7 | 66 |
Total number of gains: 98; Total number of losses: 25; Total Number of normals: 365.
Somatic mutations of KRT9:
Generating mutation plots.
Highly correlated genes for KRT9:
Showing top 20/126 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KRT9 | GPRIN1 | 0.724668 | 3 | 0 | 3 |
| KRT9 | ZSWIM3 | 0.719664 | 3 | 0 | 3 |
| KRT9 | TMEM102 | 0.715807 | 4 | 0 | 4 |
| KRT9 | REM2 | 0.710568 | 3 | 0 | 3 |
| KRT9 | HIRIP3 | 0.688391 | 3 | 0 | 3 |
| KRT9 | ALDH16A1 | 0.688212 | 3 | 0 | 3 |
| KRT9 | DLK2 | 0.679979 | 3 | 0 | 3 |
| KRT9 | NPPA | 0.670655 | 3 | 0 | 3 |
| KRT9 | BEND4 | 0.666116 | 3 | 0 | 3 |
| KRT9 | CCDC88B | 0.654275 | 3 | 0 | 3 |
| KRT9 | CACNA1A | 0.653786 | 4 | 0 | 4 |
| KRT9 | RTEL1 | 0.653155 | 3 | 0 | 3 |
| KRT9 | LIPE | 0.650508 | 3 | 0 | 3 |
| KRT9 | GNB3 | 0.650378 | 4 | 0 | 3 |
| KRT9 | CELF4 | 0.646985 | 3 | 0 | 3 |
| KRT9 | TSPAN10 | 0.645364 | 4 | 0 | 3 |
| KRT9 | PHEX | 0.645017 | 3 | 0 | 3 |
| KRT9 | ARL4D | 0.643515 | 3 | 0 | 3 |
| KRT9 | SP8 | 0.642861 | 4 | 0 | 4 |
| KRT9 | FAM207A | 0.642124 | 3 | 0 | 3 |
For details and further investigation, click here