| Full name: cancer susceptibility 2 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 10q26.11 | ||
| Entrez ID: 255082 | HGNC ID: HGNC:22933 | Ensembl Gene: ENSG00000177640 | OMIM ID: 608598 |
| Drug and gene relationship at DGIdb | |||
Expression of CASC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CASC2 | 255082 | 1562336_at | 0.0410 | 0.8819 | |
| GSE26886 | CASC2 | 255082 | 1556630_at | -0.0103 | 0.9424 | |
| GSE45670 | CASC2 | 255082 | 1562336_at | 0.0032 | 0.9803 | |
| GSE53622 | CASC2 | 255082 | 77497 | 0.2034 | 0.0291 | |
| GSE53624 | CASC2 | 255082 | 77497 | 0.1757 | 0.1403 | |
| GSE63941 | CASC2 | 255082 | 1556630_at | 0.4148 | 0.2840 | |
| GSE77861 | CASC2 | 255082 | 1556630_at | -0.1907 | 0.0974 | |
| SRP064894 | CASC2 | 255082 | RNAseq | -0.4866 | 0.0274 | |
| SRP133303 | CASC2 | 255082 | RNAseq | 0.3920 | 0.2548 | |
| SRP159526 | CASC2 | 255082 | RNAseq | -0.3032 | 0.5459 | |
| SRP219564 | CASC2 | 255082 | RNAseq | -0.8128 | 0.2307 | |
| TCGA | CASC2 | 255082 | RNAseq | -0.5585 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 0.
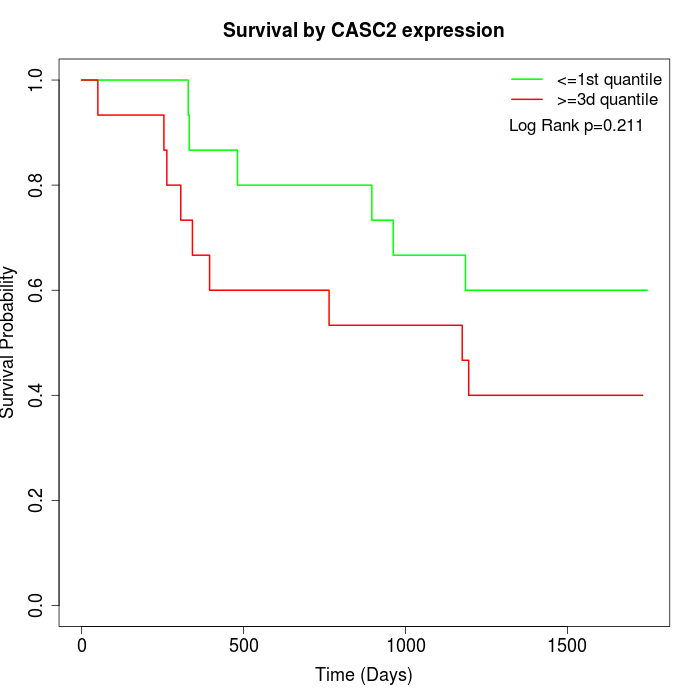
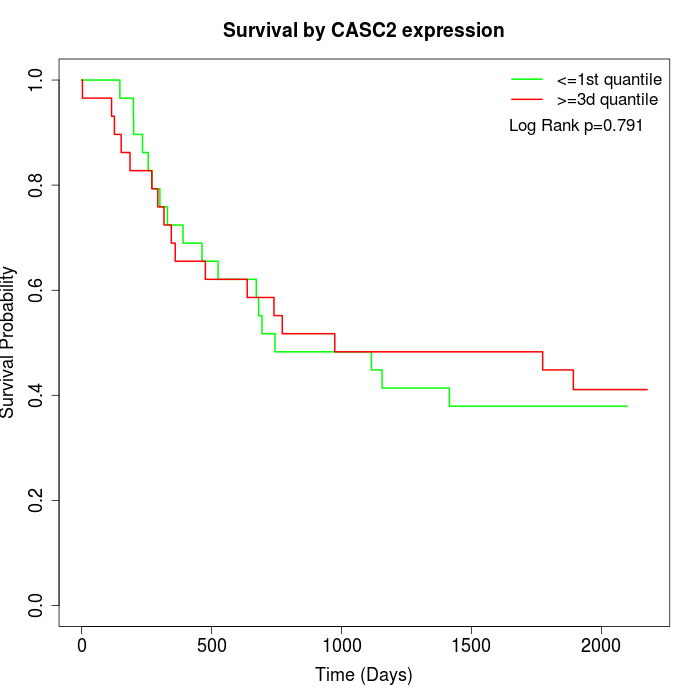
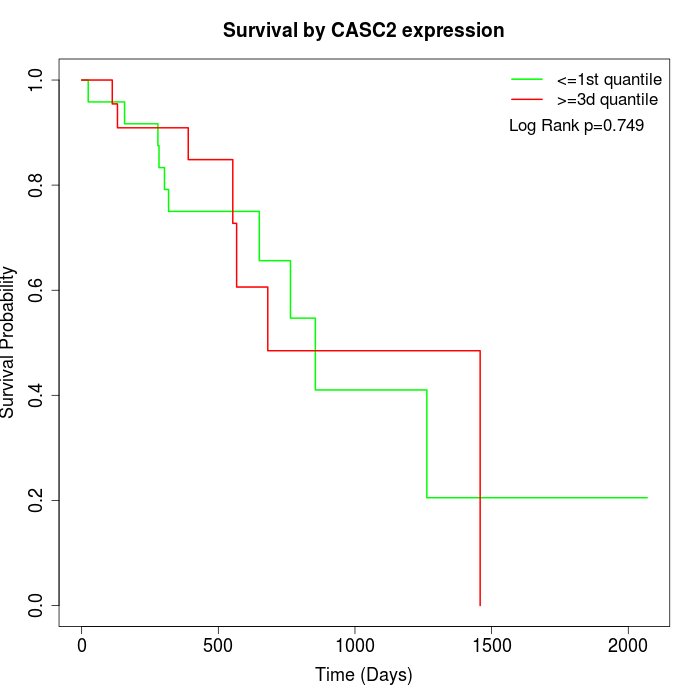
Survival by CASC2 expression:
Note: Click image to view full size file.
Copy number change of CASC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CASC2 | 255082 | 1 | 7 | 22 | |
| GSE20123 | CASC2 | 255082 | 1 | 7 | 22 | |
| GSE43470 | CASC2 | 255082 | 0 | 9 | 34 | |
| GSE46452 | CASC2 | 255082 | 0 | 11 | 48 | |
| GSE47630 | CASC2 | 255082 | 2 | 14 | 24 | |
| GSE54993 | CASC2 | 255082 | 8 | 0 | 62 | |
| GSE54994 | CASC2 | 255082 | 1 | 9 | 43 | |
| GSE60625 | CASC2 | 255082 | 0 | 0 | 11 | |
| GSE74703 | CASC2 | 255082 | 0 | 6 | 30 | |
| GSE74704 | CASC2 | 255082 | 1 | 3 | 16 | |
| TCGA | CASC2 | 255082 | 4 | 28 | 64 |
Total number of gains: 18; Total number of losses: 94; Total Number of normals: 376.
Somatic mutations of CASC2:
Generating mutation plots.
Highly correlated genes for CASC2:
Showing top 20/229 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CASC2 | COL11A2 | 0.735554 | 4 | 0 | 4 |
| CASC2 | GPR182 | 0.726309 | 5 | 0 | 5 |
| CASC2 | DSCAM-AS1 | 0.719241 | 3 | 0 | 3 |
| CASC2 | BFSP2-AS1 | 0.718898 | 3 | 0 | 3 |
| CASC2 | KRTAP2-1 | 0.718595 | 3 | 0 | 3 |
| CASC2 | BTNL2 | 0.705845 | 3 | 0 | 3 |
| CASC2 | GFRA4 | 0.702138 | 3 | 0 | 3 |
| CASC2 | ARX | 0.701533 | 4 | 0 | 3 |
| CASC2 | SLC25A53 | 0.699898 | 3 | 0 | 3 |
| CASC2 | DEFB128 | 0.696498 | 3 | 0 | 3 |
| CASC2 | CKM | 0.687504 | 3 | 0 | 3 |
| CASC2 | LINC01122 | 0.685929 | 4 | 0 | 4 |
| CASC2 | FAM222A-AS1 | 0.682234 | 4 | 0 | 4 |
| CASC2 | ZBTB46-AS1 | 0.679945 | 3 | 0 | 3 |
| CASC2 | LIPE-AS1 | 0.679918 | 3 | 0 | 3 |
| CASC2 | KCNJ9 | 0.675013 | 3 | 0 | 3 |
| CASC2 | SLC4A8 | 0.670657 | 3 | 0 | 3 |
| CASC2 | HGFAC | 0.670587 | 3 | 0 | 3 |
| CASC2 | SLC22A12 | 0.668677 | 3 | 0 | 3 |
| CASC2 | GRIA4 | 0.668515 | 3 | 0 | 3 |
For details and further investigation, click here