| Full name: brain expressed X-linked 4 | Alias Symbol: FLJ10097 | ||
| Type: protein-coding gene | Cytoband: Xq22.1 | ||
| Entrez ID: 56271 | HGNC ID: HGNC:25475 | Ensembl Gene: ENSG00000102409 | OMIM ID: 300692 |
| Drug and gene relationship at DGIdb | |||
Expression of BEX4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BEX4 | 56271 | 215440_s_at | -0.9451 | 0.0596 | |
| GSE20347 | BEX4 | 56271 | 215440_s_at | -3.2134 | 0.0000 | |
| GSE23400 | BEX4 | 56271 | 215440_s_at | -1.6324 | 0.0000 | |
| GSE26886 | BEX4 | 56271 | 215440_s_at | -3.0803 | 0.0000 | |
| GSE29001 | BEX4 | 56271 | 215440_s_at | -2.6336 | 0.0034 | |
| GSE38129 | BEX4 | 56271 | 215440_s_at | -2.3516 | 0.0000 | |
| GSE45670 | BEX4 | 56271 | 215440_s_at | -1.6141 | 0.0002 | |
| GSE53622 | BEX4 | 56271 | 91900 | -1.7770 | 0.0000 | |
| GSE53624 | BEX4 | 56271 | 91900 | -2.0539 | 0.0000 | |
| GSE63941 | BEX4 | 56271 | 215440_s_at | -3.0746 | 0.1285 | |
| GSE77861 | BEX4 | 56271 | 215440_s_at | -3.0559 | 0.0016 | |
| GSE97050 | BEX4 | 56271 | A_24_P40551 | -1.4181 | 0.0620 | |
| SRP007169 | BEX4 | 56271 | RNAseq | -4.0305 | 0.0000 | |
| SRP008496 | BEX4 | 56271 | RNAseq | -3.1237 | 0.0000 | |
| SRP064894 | BEX4 | 56271 | RNAseq | -1.5881 | 0.0000 | |
| SRP133303 | BEX4 | 56271 | RNAseq | -1.4670 | 0.0000 | |
| SRP159526 | BEX4 | 56271 | RNAseq | -0.5666 | 0.1434 | |
| SRP193095 | BEX4 | 56271 | RNAseq | -1.7131 | 0.0000 | |
| SRP219564 | BEX4 | 56271 | RNAseq | -1.4713 | 0.0053 | |
| TCGA | BEX4 | 56271 | RNAseq | -0.3436 | 0.0287 |
Upregulated datasets: 0; Downregulated datasets: 15.
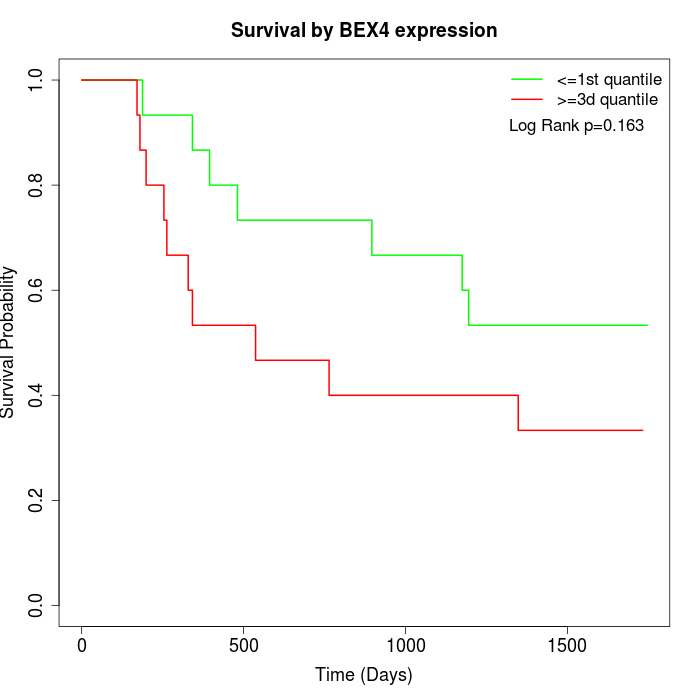
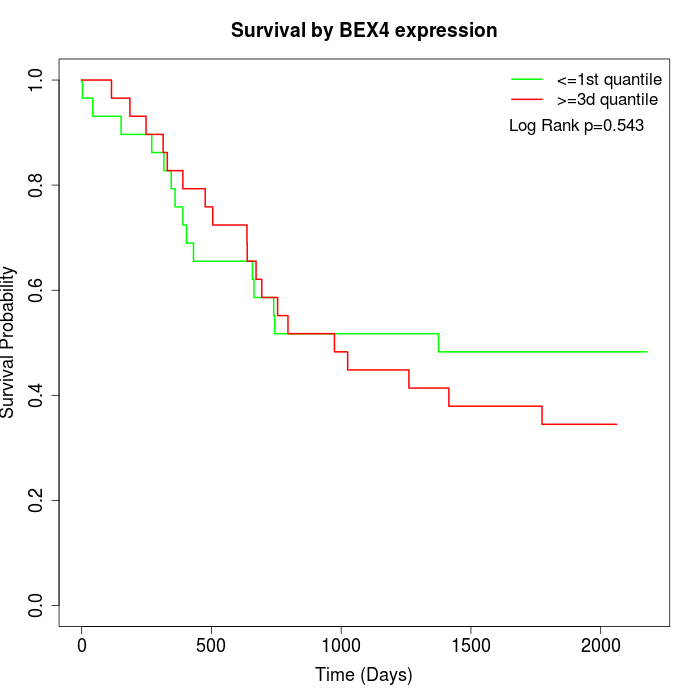
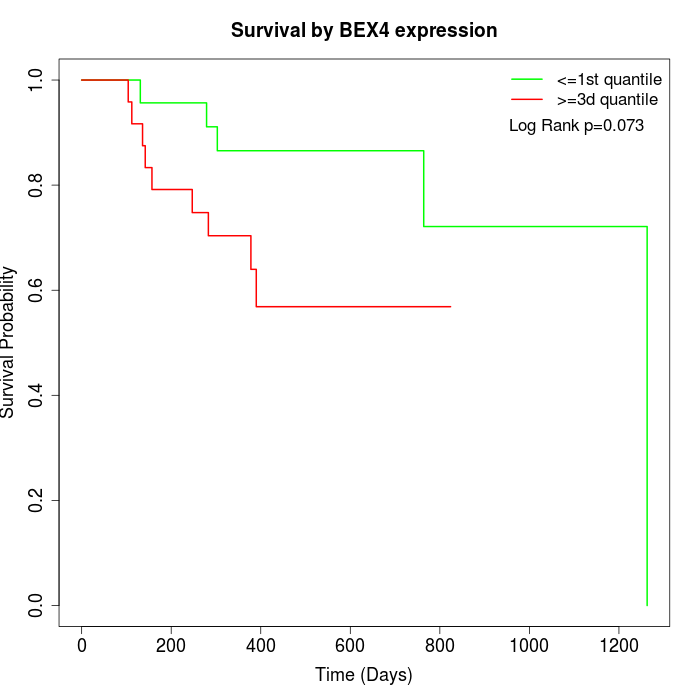
Survival by BEX4 expression:
Note: Click image to view full size file.
Copy number change of BEX4:
No record found for this gene.
Somatic mutations of BEX4:
Generating mutation plots.
Highly correlated genes for BEX4:
Showing top 20/1729 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BEX4 | SH3BGRL2 | 0.799316 | 7 | 0 | 7 |
| BEX4 | GPD1L | 0.785301 | 11 | 0 | 11 |
| BEX4 | LMBRD1 | 0.784692 | 11 | 0 | 11 |
| BEX4 | SHROOM3 | 0.781382 | 7 | 0 | 7 |
| BEX4 | CCIN | 0.777263 | 4 | 0 | 4 |
| BEX4 | HPGD | 0.772808 | 11 | 0 | 11 |
| BEX4 | UBL3 | 0.772482 | 10 | 0 | 10 |
| BEX4 | RNF103 | 0.761946 | 11 | 0 | 11 |
| BEX4 | NCOA1 | 0.76056 | 10 | 0 | 10 |
| BEX4 | ATF7 | 0.758555 | 3 | 0 | 3 |
| BEX4 | MTMR10 | 0.758096 | 7 | 0 | 7 |
| BEX4 | LSM11 | 0.757462 | 3 | 0 | 3 |
| BEX4 | PAIP2B | 0.754702 | 11 | 0 | 11 |
| BEX4 | ID4 | 0.75443 | 9 | 0 | 9 |
| BEX4 | NAA35 | 0.751789 | 3 | 0 | 3 |
| BEX4 | ECHDC2 | 0.750762 | 11 | 0 | 11 |
| BEX4 | RAB5B | 0.75036 | 10 | 0 | 10 |
| BEX4 | RBM20 | 0.750351 | 4 | 0 | 4 |
| BEX4 | MPP7 | 0.750297 | 7 | 0 | 7 |
| BEX4 | ELMO2 | 0.748117 | 11 | 0 | 11 |
For details and further investigation, click here