| Full name: chaperonin containing TCP1 subunit 8 like 2 | Alias Symbol: CESK1 | ||
| Type: protein-coding gene | Cytoband: 22q11.1 | ||
| Entrez ID: 150160 | HGNC ID: HGNC:15553 | Ensembl Gene: ENSG00000198445 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CCT8L2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCT8L2 | 150160 | 220508_at | 0.1329 | 0.5476 | |
| GSE20347 | CCT8L2 | 150160 | 220508_at | 0.0742 | 0.4180 | |
| GSE23400 | CCT8L2 | 150160 | 220508_at | -0.0913 | 0.0064 | |
| GSE26886 | CCT8L2 | 150160 | 220508_at | 0.0913 | 0.4629 | |
| GSE29001 | CCT8L2 | 150160 | 220508_at | -0.1937 | 0.0718 | |
| GSE38129 | CCT8L2 | 150160 | 220508_at | -0.0804 | 0.2353 | |
| GSE45670 | CCT8L2 | 150160 | 220508_at | -0.1311 | 0.1520 | |
| GSE53622 | CCT8L2 | 150160 | 42082 | 0.1202 | 0.1914 | |
| GSE53624 | CCT8L2 | 150160 | 42082 | 0.0097 | 0.9180 | |
| GSE63941 | CCT8L2 | 150160 | 220508_at | 0.0694 | 0.6623 | |
| GSE77861 | CCT8L2 | 150160 | 220508_at | -0.1902 | 0.3417 | |
| TCGA | CCT8L2 | 150160 | RNAseq | 0.1338 | 0.9425 |
Upregulated datasets: 0; Downregulated datasets: 0.
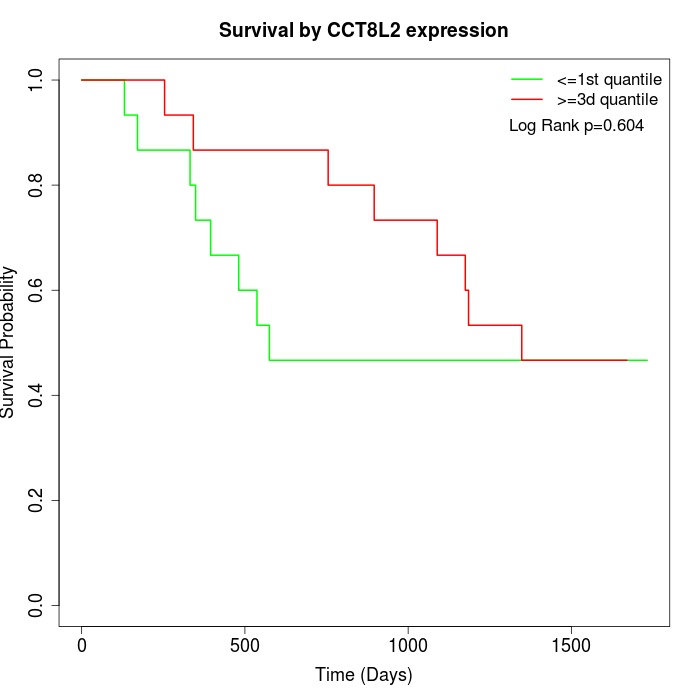
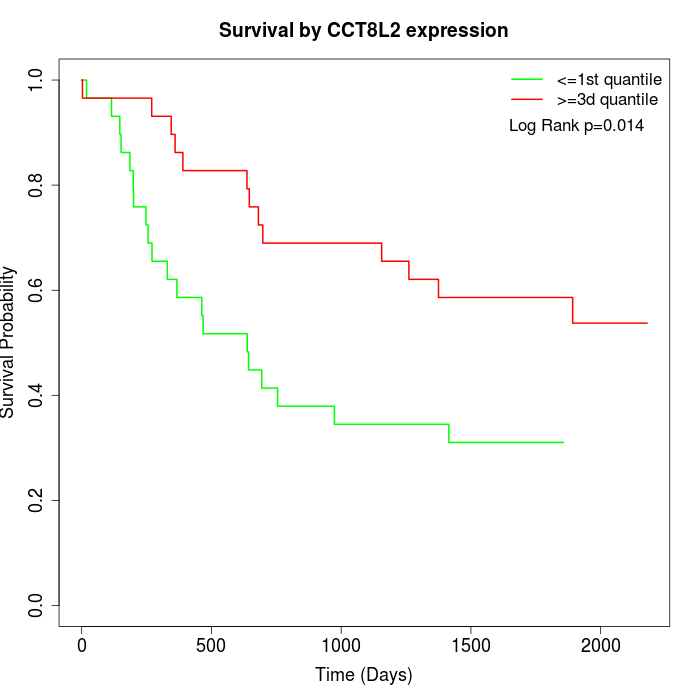
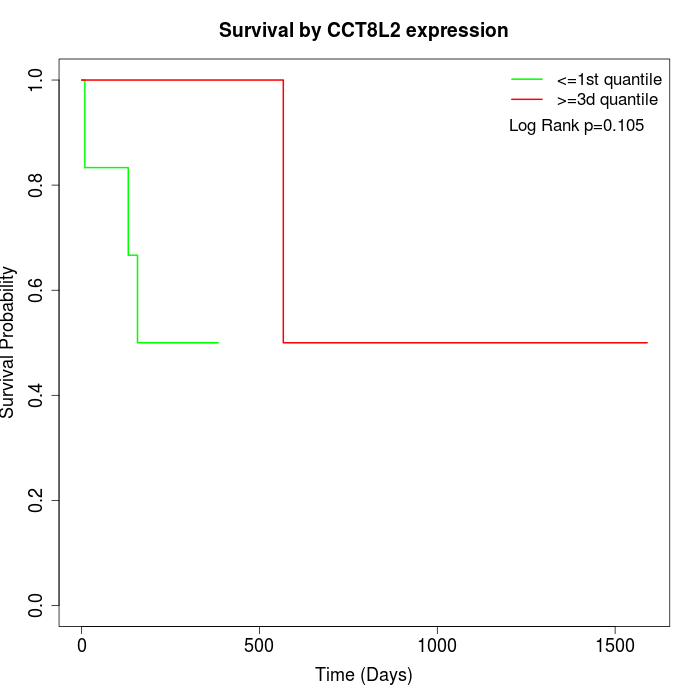
Survival by CCT8L2 expression:
Note: Click image to view full size file.
Copy number change of CCT8L2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCT8L2 | 150160 | 3 | 8 | 19 | |
| GSE20123 | CCT8L2 | 150160 | 3 | 8 | 19 | |
| GSE43470 | CCT8L2 | 150160 | 3 | 8 | 32 | |
| GSE46452 | CCT8L2 | 150160 | 32 | 2 | 25 | |
| GSE47630 | CCT8L2 | 150160 | 8 | 5 | 27 | |
| GSE54993 | CCT8L2 | 150160 | 2 | 7 | 61 | |
| GSE54994 | CCT8L2 | 150160 | 11 | 7 | 35 | |
| GSE60625 | CCT8L2 | 150160 | 5 | 0 | 6 | |
| GSE74703 | CCT8L2 | 150160 | 3 | 6 | 27 | |
| GSE74704 | CCT8L2 | 150160 | 1 | 5 | 14 | |
| TCGA | CCT8L2 | 150160 | 25 | 17 | 54 |
Total number of gains: 96; Total number of losses: 73; Total Number of normals: 319.
Somatic mutations of CCT8L2:
Generating mutation plots.
Highly correlated genes for CCT8L2:
Showing top 20/590 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCT8L2 | BTBD17 | 0.68414 | 4 | 0 | 4 |
| CCT8L2 | SPSB4 | 0.675913 | 3 | 0 | 3 |
| CCT8L2 | ANKRD60 | 0.675249 | 3 | 0 | 3 |
| CCT8L2 | TTC9B | 0.667597 | 3 | 0 | 3 |
| CCT8L2 | EDA2R | 0.66229 | 4 | 0 | 4 |
| CCT8L2 | PHOX2B | 0.657667 | 4 | 0 | 3 |
| CCT8L2 | OVGP1 | 0.650967 | 3 | 0 | 3 |
| CCT8L2 | CTRB2 | 0.646626 | 7 | 0 | 7 |
| CCT8L2 | LILRB5 | 0.638974 | 4 | 0 | 3 |
| CCT8L2 | SPATA3 | 0.638488 | 4 | 0 | 4 |
| CCT8L2 | CNIH2 | 0.632218 | 3 | 0 | 3 |
| CCT8L2 | ASCL1 | 0.63092 | 4 | 0 | 3 |
| CCT8L2 | AMN | 0.622795 | 7 | 0 | 7 |
| CCT8L2 | CHRNA2 | 0.621242 | 6 | 0 | 6 |
| CCT8L2 | ANK1 | 0.620757 | 5 | 0 | 4 |
| CCT8L2 | TBX5 | 0.618914 | 4 | 0 | 4 |
| CCT8L2 | C11orf86 | 0.618257 | 3 | 0 | 3 |
| CCT8L2 | SPAG6 | 0.618045 | 5 | 0 | 4 |
| CCT8L2 | SLC30A3 | 0.615605 | 5 | 0 | 4 |
| CCT8L2 | GJA3 | 0.614776 | 4 | 0 | 3 |
For details and further investigation, click here