| Full name: BTB domain containing 17 | Alias Symbol: LGALS3BPL|BTBD17A|TANGO10A | ||
| Type: protein-coding gene | Cytoband: 17q25.1 | ||
| Entrez ID: 388419 | HGNC ID: HGNC:33758 | Ensembl Gene: ENSG00000204347 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of BTBD17:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BTBD17 | 388419 | 230445_at | 0.0803 | 0.7312 | |
| GSE26886 | BTBD17 | 388419 | 230445_at | 0.2458 | 0.0339 | |
| GSE45670 | BTBD17 | 388419 | 230445_at | -0.0156 | 0.9054 | |
| GSE53622 | BTBD17 | 388419 | 10499 | -0.1275 | 0.2228 | |
| GSE53624 | BTBD17 | 388419 | 10499 | 0.0774 | 0.6217 | |
| GSE63941 | BTBD17 | 388419 | 230445_at | 0.1991 | 0.2219 | |
| GSE77861 | BTBD17 | 388419 | 230445_at | -0.1133 | 0.4758 | |
| TCGA | BTBD17 | 388419 | RNAseq | -1.5060 | 0.1745 |
Upregulated datasets: 0; Downregulated datasets: 0.
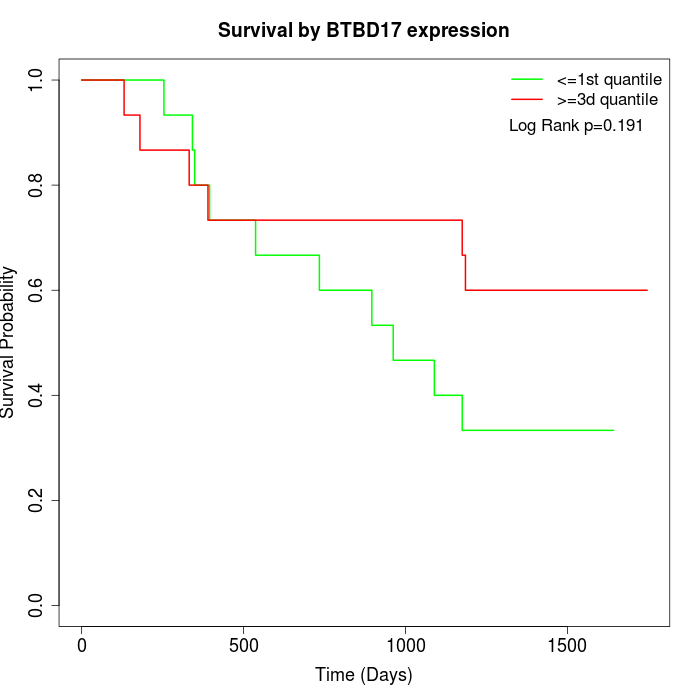
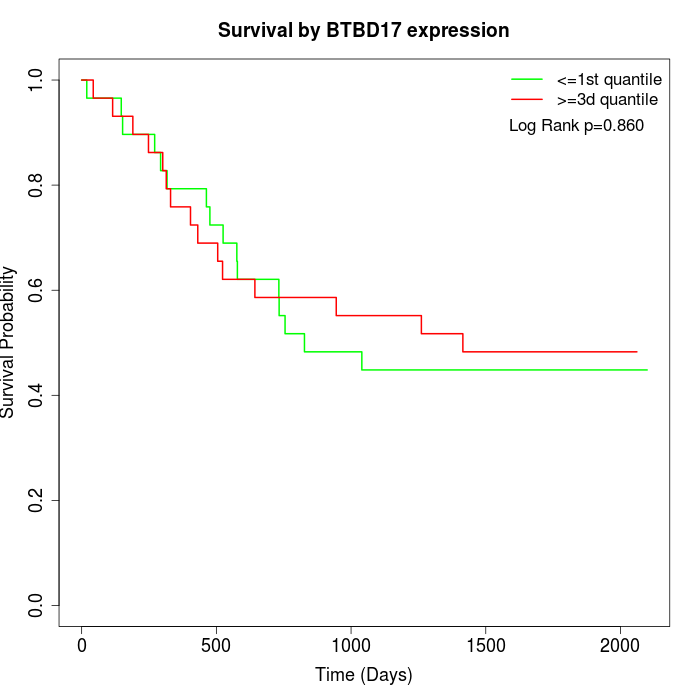
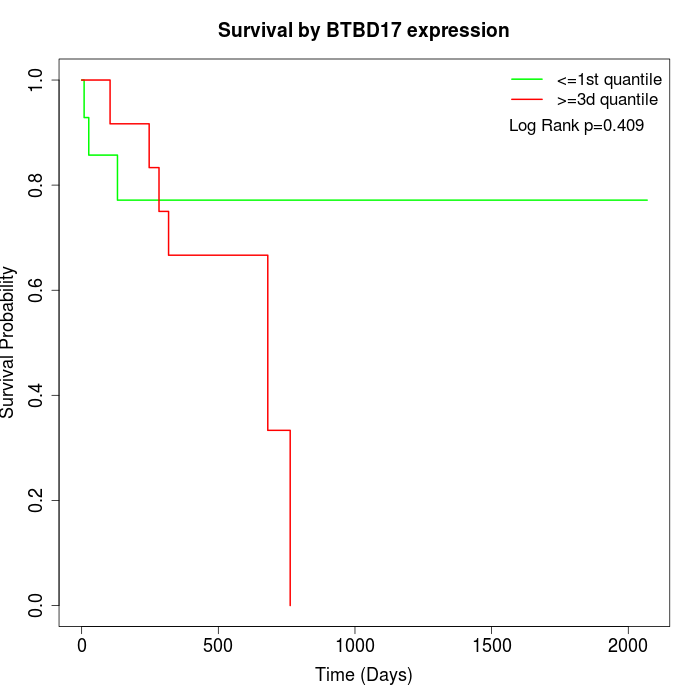
Survival by BTBD17 expression:
Note: Click image to view full size file.
Copy number change of BTBD17:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BTBD17 | 388419 | 3 | 2 | 25 | |
| GSE20123 | BTBD17 | 388419 | 3 | 2 | 25 | |
| GSE43470 | BTBD17 | 388419 | 5 | 3 | 35 | |
| GSE46452 | BTBD17 | 388419 | 33 | 0 | 26 | |
| GSE47630 | BTBD17 | 388419 | 8 | 1 | 31 | |
| GSE54993 | BTBD17 | 388419 | 2 | 5 | 63 | |
| GSE54994 | BTBD17 | 388419 | 10 | 4 | 39 | |
| GSE60625 | BTBD17 | 388419 | 6 | 0 | 5 | |
| GSE74703 | BTBD17 | 388419 | 5 | 1 | 30 | |
| GSE74704 | BTBD17 | 388419 | 3 | 1 | 16 | |
| TCGA | BTBD17 | 388419 | 32 | 10 | 54 |
Total number of gains: 110; Total number of losses: 29; Total Number of normals: 349.
Somatic mutations of BTBD17:
Generating mutation plots.
Highly correlated genes for BTBD17:
Showing top 20/99 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BTBD17 | AFMID | 0.705989 | 3 | 0 | 3 |
| BTBD17 | SHOX2 | 0.700152 | 3 | 0 | 3 |
| BTBD17 | CATSPER3 | 0.691682 | 3 | 0 | 3 |
| BTBD17 | CCT8L2 | 0.68414 | 4 | 0 | 4 |
| BTBD17 | PKLR | 0.668029 | 3 | 0 | 3 |
| BTBD17 | LINC01169 | 0.660386 | 3 | 0 | 3 |
| BTBD17 | PLD4 | 0.658246 | 3 | 0 | 3 |
| BTBD17 | CLEC4M | 0.654347 | 4 | 0 | 3 |
| BTBD17 | NTSR1 | 0.648329 | 3 | 0 | 3 |
| BTBD17 | ITGA2B | 0.646367 | 3 | 0 | 3 |
| BTBD17 | LINC00311 | 0.645699 | 3 | 0 | 3 |
| BTBD17 | ACHE | 0.643365 | 3 | 0 | 3 |
| BTBD17 | SLC8A3 | 0.641661 | 3 | 0 | 3 |
| BTBD17 | FCGR1A | 0.638503 | 3 | 0 | 3 |
| BTBD17 | SIX6 | 0.630014 | 4 | 0 | 4 |
| BTBD17 | SYP | 0.625109 | 3 | 0 | 3 |
| BTBD17 | HHIP | 0.625072 | 3 | 0 | 3 |
| BTBD17 | XIRP2 | 0.624752 | 3 | 0 | 3 |
| BTBD17 | CCR10 | 0.623642 | 4 | 0 | 3 |
| BTBD17 | SNX8 | 0.6211 | 3 | 0 | 3 |
For details and further investigation, click here