| Full name: chymotrypsinogen B2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16q23.1 | ||
| Entrez ID: 440387 | HGNC ID: HGNC:2522 | Ensembl Gene: ENSG00000168928 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CTRB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CTRB2 | 440387 | 214411_x_at | 0.0683 | 0.7927 | |
| GSE20347 | CTRB2 | 440387 | 214411_x_at | 0.0094 | 0.9231 | |
| GSE23400 | CTRB2 | 440387 | 214411_x_at | -0.0857 | 0.1714 | |
| GSE26886 | CTRB2 | 440387 | 214411_x_at | 0.1116 | 0.3018 | |
| GSE29001 | CTRB2 | 440387 | 214411_x_at | -0.1074 | 0.3588 | |
| GSE38129 | CTRB2 | 440387 | 214411_x_at | -0.0195 | 0.8151 | |
| GSE45670 | CTRB2 | 440387 | 214411_x_at | 0.1647 | 0.0294 | |
| GSE53622 | CTRB2 | 440387 | 92980 | 0.2234 | 0.0005 | |
| GSE53624 | CTRB2 | 440387 | 92980 | -0.0752 | 0.3597 | |
| GSE63941 | CTRB2 | 440387 | 214411_x_at | 0.1588 | 0.2410 | |
| GSE77861 | CTRB2 | 440387 | 214411_x_at | -0.0384 | 0.7497 | |
| GSE97050 | CTRB2 | 440387 | A_32_P86150 | 0.3942 | 0.1403 | |
| TCGA | CTRB2 | 440387 | RNAseq | 1.0149 | 0.6152 |
Upregulated datasets: 0; Downregulated datasets: 0.
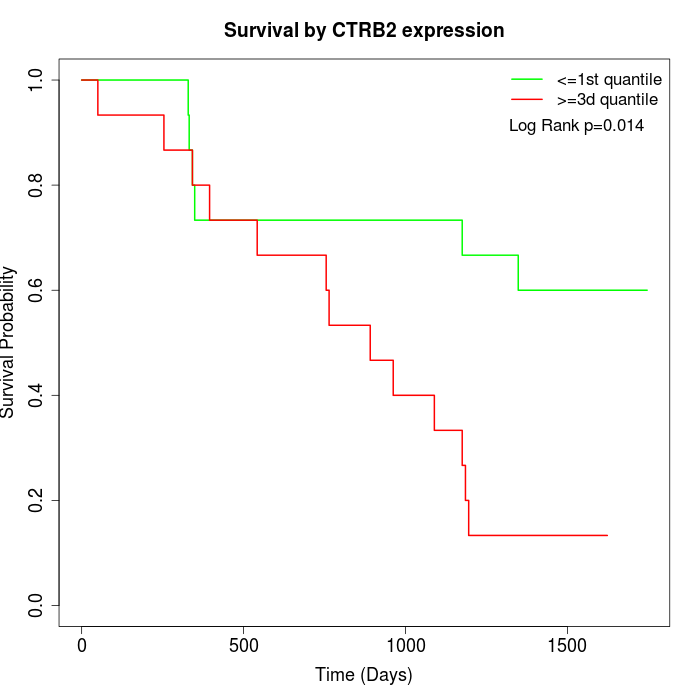
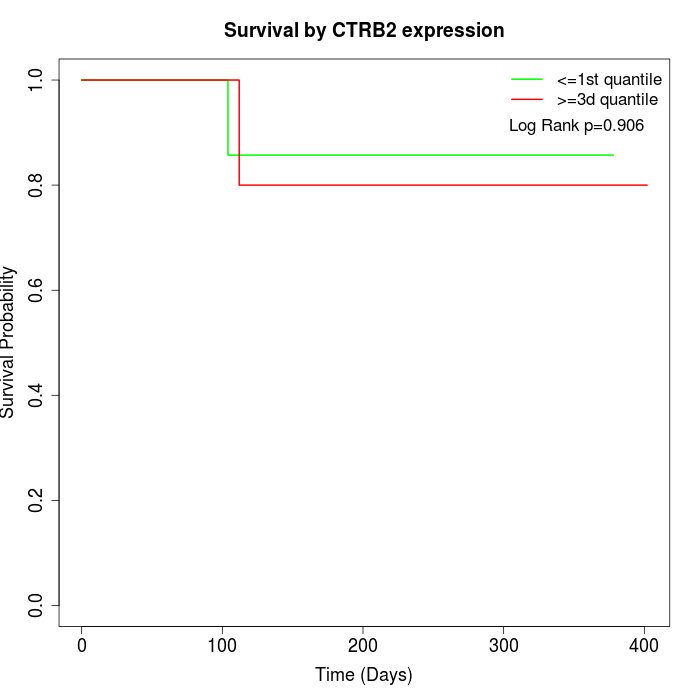
Survival by CTRB2 expression:
Note: Click image to view full size file.
Copy number change of CTRB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTRB2 | 440387 | 6 | 1 | 23 | |
| GSE20123 | CTRB2 | 440387 | 6 | 1 | 23 | |
| GSE43470 | CTRB2 | 440387 | 2 | 8 | 33 | |
| GSE46452 | CTRB2 | 440387 | 37 | 2 | 20 | |
| GSE47630 | CTRB2 | 440387 | 11 | 8 | 21 | |
| GSE54993 | CTRB2 | 440387 | 2 | 4 | 64 | |
| GSE54994 | CTRB2 | 440387 | 8 | 10 | 35 | |
| GSE60625 | CTRB2 | 440387 | 4 | 0 | 7 | |
| GSE74703 | CTRB2 | 440387 | 2 | 5 | 29 | |
| GSE74704 | CTRB2 | 440387 | 3 | 0 | 17 | |
| TCGA | CTRB2 | 440387 | 25 | 15 | 56 |
Total number of gains: 106; Total number of losses: 54; Total Number of normals: 328.
Somatic mutations of CTRB2:
Generating mutation plots.
Highly correlated genes for CTRB2:
Showing top 20/714 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTRB2 | ICAM1 | 0.728794 | 3 | 0 | 3 |
| CTRB2 | ANPEP | 0.723909 | 3 | 0 | 3 |
| CTRB2 | RNF31 | 0.71225 | 3 | 0 | 3 |
| CTRB2 | E2F4 | 0.707301 | 6 | 0 | 6 |
| CTRB2 | KIR3DL2 | 0.701837 | 3 | 0 | 3 |
| CTRB2 | PDE4A | 0.700157 | 3 | 0 | 3 |
| CTRB2 | PPP1R18 | 0.693301 | 3 | 0 | 3 |
| CTRB2 | RGSL1 | 0.690227 | 3 | 0 | 3 |
| CTRB2 | ITGB3 | 0.690135 | 7 | 0 | 6 |
| CTRB2 | ZYX | 0.687797 | 3 | 0 | 3 |
| CTRB2 | TDO2 | 0.687662 | 3 | 0 | 3 |
| CTRB2 | OR2T12 | 0.684213 | 3 | 0 | 3 |
| CTRB2 | CCL22 | 0.683346 | 4 | 0 | 4 |
| CTRB2 | LMF2 | 0.682963 | 4 | 0 | 3 |
| CTRB2 | ELFN1 | 0.681868 | 5 | 0 | 4 |
| CTRB2 | ZNF358 | 0.681626 | 3 | 0 | 3 |
| CTRB2 | CHRM5 | 0.681031 | 5 | 0 | 5 |
| CTRB2 | ARHGEF33 | 0.680749 | 4 | 0 | 4 |
| CTRB2 | LMAN1 | 0.676255 | 3 | 0 | 3 |
| CTRB2 | PSAPL1 | 0.6736 | 3 | 0 | 3 |
For details and further investigation, click here