| Full name: chloride intracellular channel 2 | Alias Symbol: XAP121|CLCNL2 | ||
| Type: protein-coding gene | Cytoband: Xq28 | ||
| Entrez ID: 1193 | HGNC ID: HGNC:2063 | Ensembl Gene: ENSG00000155962 | OMIM ID: 300138 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CLIC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLIC2 | 1193 | 213415_at | 0.0871 | 0.9363 | |
| GSE20347 | CLIC2 | 1193 | 213415_at | 0.7577 | 0.0508 | |
| GSE23400 | CLIC2 | 1193 | 213415_at | 0.0378 | 0.6636 | |
| GSE26886 | CLIC2 | 1193 | 213415_at | -0.0176 | 0.9738 | |
| GSE29001 | CLIC2 | 1193 | 213415_at | 0.1878 | 0.6155 | |
| GSE38129 | CLIC2 | 1193 | 213415_at | 0.2750 | 0.4915 | |
| GSE45670 | CLIC2 | 1193 | 213415_at | -0.1371 | 0.7738 | |
| GSE53622 | CLIC2 | 1193 | 34752 | -0.3042 | 0.0636 | |
| GSE53624 | CLIC2 | 1193 | 34752 | -0.1063 | 0.4792 | |
| GSE63941 | CLIC2 | 1193 | 213415_at | -1.2183 | 0.2922 | |
| GSE77861 | CLIC2 | 1193 | 213415_at | 0.5472 | 0.2478 | |
| GSE97050 | CLIC2 | 1193 | A_33_P3775848 | -0.0033 | 0.9916 | |
| SRP007169 | CLIC2 | 1193 | RNAseq | -0.8369 | 0.1203 | |
| SRP008496 | CLIC2 | 1193 | RNAseq | -1.3011 | 0.0021 | |
| SRP064894 | CLIC2 | 1193 | RNAseq | -0.1090 | 0.6982 | |
| SRP133303 | CLIC2 | 1193 | RNAseq | -0.1831 | 0.4891 | |
| SRP159526 | CLIC2 | 1193 | RNAseq | 0.0287 | 0.9263 | |
| SRP193095 | CLIC2 | 1193 | RNAseq | 0.2989 | 0.1063 | |
| SRP219564 | CLIC2 | 1193 | RNAseq | 0.2182 | 0.6019 | |
| TCGA | CLIC2 | 1193 | RNAseq | -0.0441 | 0.7521 |
Upregulated datasets: 0; Downregulated datasets: 1.
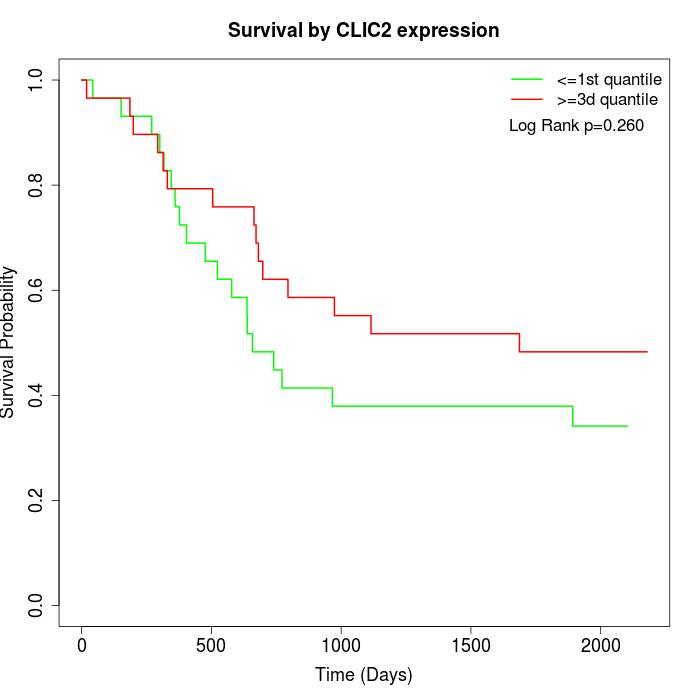
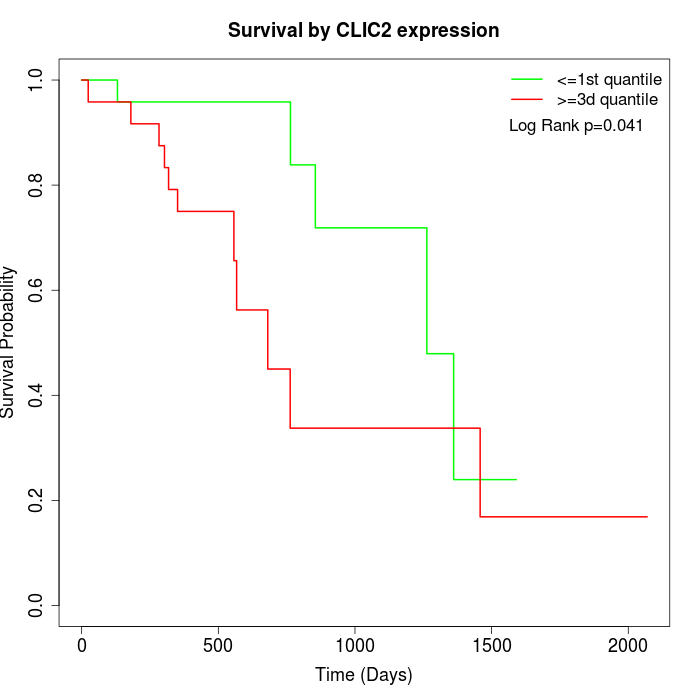
Survival by CLIC2 expression:
Note: Click image to view full size file.
Copy number change of CLIC2:
No record found for this gene.
Somatic mutations of CLIC2:
Generating mutation plots.
Highly correlated genes for CLIC2:
Showing top 20/298 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLIC2 | IL6R | 0.79819 | 3 | 0 | 3 |
| CLIC2 | CHN1 | 0.736433 | 3 | 0 | 3 |
| CLIC2 | CHD1 | 0.725567 | 3 | 0 | 3 |
| CLIC2 | CDKN1C | 0.724633 | 3 | 0 | 3 |
| CLIC2 | GLIPR1 | 0.716929 | 3 | 0 | 3 |
| CLIC2 | TTYH2 | 0.713475 | 3 | 0 | 3 |
| CLIC2 | TNFAIP8L2 | 0.708092 | 3 | 0 | 3 |
| CLIC2 | UGGT2 | 0.68365 | 5 | 0 | 5 |
| CLIC2 | PPIG | 0.682465 | 3 | 0 | 3 |
| CLIC2 | LILRB5 | 0.678432 | 5 | 0 | 4 |
| CLIC2 | C19orf38 | 0.675937 | 3 | 0 | 3 |
| CLIC2 | ANKRD10 | 0.674815 | 4 | 0 | 3 |
| CLIC2 | KRTCAP2 | 0.674227 | 3 | 0 | 3 |
| CLIC2 | DEGS1 | 0.671841 | 4 | 0 | 3 |
| CLIC2 | ARHGAP15 | 0.667765 | 4 | 0 | 4 |
| CLIC2 | ZC3H15 | 0.665224 | 3 | 0 | 3 |
| CLIC2 | CD99L2 | 0.662754 | 3 | 0 | 3 |
| CLIC2 | GIMAP5 | 0.6612 | 4 | 0 | 3 |
| CLIC2 | SAMD3 | 0.659966 | 4 | 0 | 3 |
| CLIC2 | ECSCR | 0.655651 | 5 | 0 | 3 |
For details and further investigation, click here