| Full name: sterile alpha motif domain containing 3 | Alias Symbol: bA73O6.2|FLJ34032 | ||
| Type: protein-coding gene | Cytoband: 6q23.1 | ||
| Entrez ID: 154075 | HGNC ID: HGNC:21574 | Ensembl Gene: ENSG00000164483 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SAMD3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SAMD3 | 154075 | 236782_at | 0.1362 | 0.8550 | |
| GSE26886 | SAMD3 | 154075 | 236782_at | -0.1338 | 0.3618 | |
| GSE45670 | SAMD3 | 154075 | 236782_at | -0.2746 | 0.1348 | |
| GSE53622 | SAMD3 | 154075 | 31846 | -0.2497 | 0.2488 | |
| GSE53624 | SAMD3 | 154075 | 31846 | 0.0329 | 0.8761 | |
| GSE63941 | SAMD3 | 154075 | 236782_at | -0.2331 | 0.5454 | |
| GSE77861 | SAMD3 | 154075 | 236782_at | -0.1677 | 0.0604 | |
| GSE97050 | SAMD3 | 154075 | A_23_P93524 | 0.0867 | 0.7445 | |
| SRP133303 | SAMD3 | 154075 | RNAseq | -0.5060 | 0.0767 | |
| SRP159526 | SAMD3 | 154075 | RNAseq | -0.0557 | 0.9384 | |
| TCGA | SAMD3 | 154075 | RNAseq | 0.3146 | 0.4718 |
Upregulated datasets: 0; Downregulated datasets: 0.
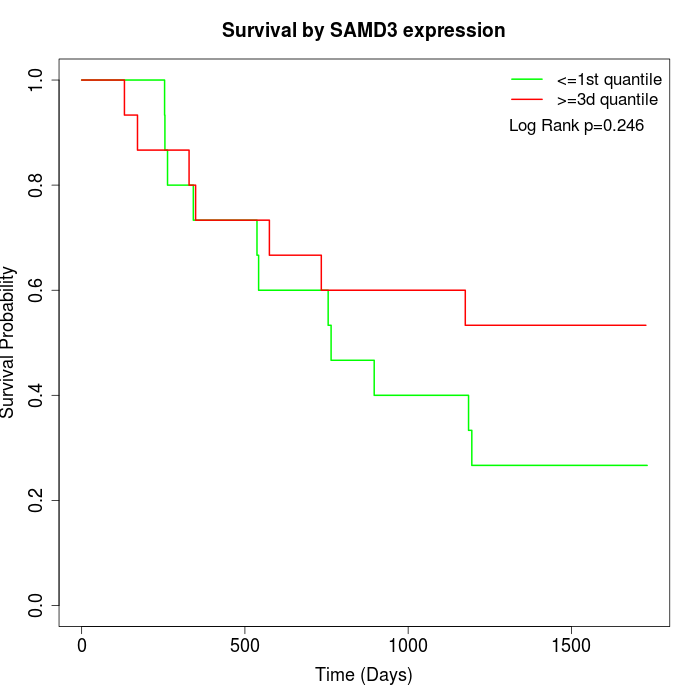
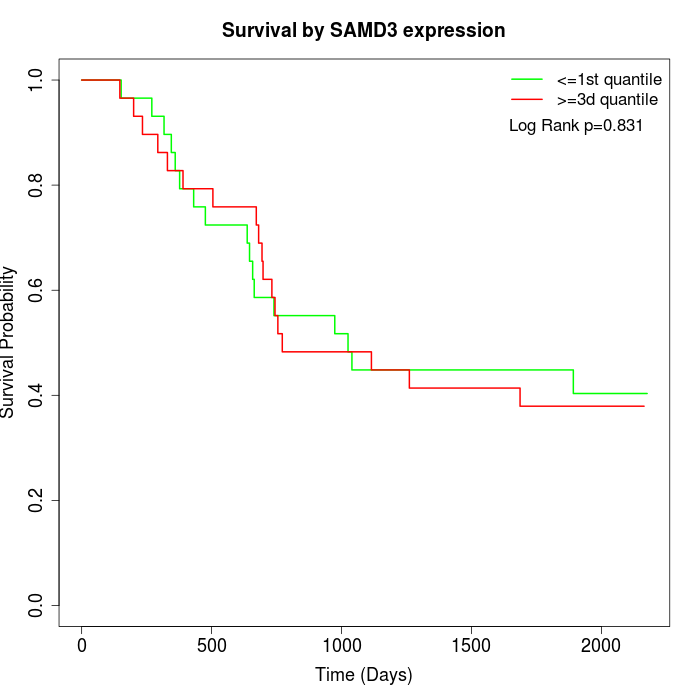
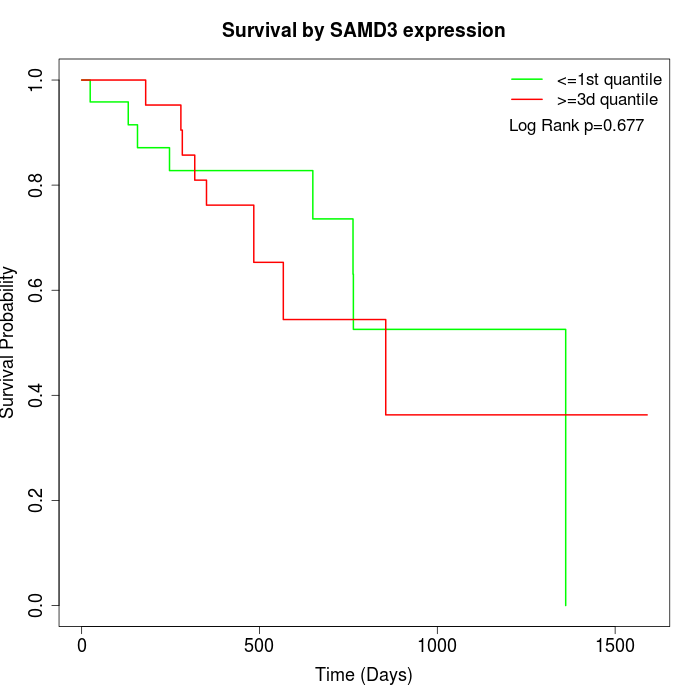
Survival by SAMD3 expression:
Note: Click image to view full size file.
Copy number change of SAMD3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SAMD3 | 154075 | 1 | 4 | 25 | |
| GSE20123 | SAMD3 | 154075 | 1 | 3 | 26 | |
| GSE43470 | SAMD3 | 154075 | 5 | 0 | 38 | |
| GSE46452 | SAMD3 | 154075 | 2 | 11 | 46 | |
| GSE47630 | SAMD3 | 154075 | 9 | 4 | 27 | |
| GSE54993 | SAMD3 | 154075 | 3 | 3 | 64 | |
| GSE54994 | SAMD3 | 154075 | 9 | 7 | 37 | |
| GSE60625 | SAMD3 | 154075 | 0 | 1 | 10 | |
| GSE74703 | SAMD3 | 154075 | 4 | 0 | 32 | |
| GSE74704 | SAMD3 | 154075 | 0 | 1 | 19 | |
| TCGA | SAMD3 | 154075 | 11 | 18 | 67 |
Total number of gains: 45; Total number of losses: 52; Total Number of normals: 391.
Somatic mutations of SAMD3:
Generating mutation plots.
Highly correlated genes for SAMD3:
Showing top 20/339 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SAMD3 | KCTD6 | 0.784773 | 3 | 0 | 3 |
| SAMD3 | BIN2 | 0.781616 | 5 | 0 | 5 |
| SAMD3 | PREX1 | 0.769444 | 3 | 0 | 3 |
| SAMD3 | CCR4 | 0.769365 | 3 | 0 | 3 |
| SAMD3 | TNFSF8 | 0.766612 | 4 | 0 | 4 |
| SAMD3 | LILRA4 | 0.763226 | 3 | 0 | 3 |
| SAMD3 | NKG7 | 0.755698 | 6 | 0 | 5 |
| SAMD3 | CCR5 | 0.75164 | 5 | 0 | 5 |
| SAMD3 | SNX20 | 0.74296 | 6 | 0 | 6 |
| SAMD3 | MADD | 0.741934 | 3 | 0 | 3 |
| SAMD3 | NLRP3 | 0.741381 | 3 | 0 | 3 |
| SAMD3 | CCL5 | 0.739946 | 5 | 0 | 5 |
| SAMD3 | UBASH3A | 0.734203 | 6 | 0 | 5 |
| SAMD3 | GPR171 | 0.731789 | 6 | 0 | 6 |
| SAMD3 | GNGT2 | 0.728634 | 6 | 0 | 6 |
| SAMD3 | GIMAP1 | 0.728624 | 5 | 0 | 5 |
| SAMD3 | CST7 | 0.728319 | 6 | 0 | 5 |
| SAMD3 | DHX30 | 0.725864 | 3 | 0 | 3 |
| SAMD3 | NCKAP1L | 0.720035 | 6 | 0 | 5 |
| SAMD3 | GIMAP5 | 0.717688 | 4 | 0 | 3 |
For details and further investigation, click here