| Full name: Rho GTPase activating protein 15 | Alias Symbol: BM046 | ||
| Type: protein-coding gene | Cytoband: 2q22.2-q22.3 | ||
| Entrez ID: 55843 | HGNC ID: HGNC:21030 | Ensembl Gene: ENSG00000075884 | OMIM ID: 610578 |
| Drug and gene relationship at DGIdb | |||
Expression of ARHGAP15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | ARHGAP15 | 55843 | 9068 | -0.7214 | 0.0000 | |
| GSE53624 | ARHGAP15 | 55843 | 9068 | -0.9135 | 0.0000 | |
| GSE97050 | ARHGAP15 | 55843 | A_23_P84154 | 0.2447 | 0.3513 | |
| SRP007169 | ARHGAP15 | 55843 | RNAseq | 0.8384 | 0.2640 | |
| SRP064894 | ARHGAP15 | 55843 | RNAseq | 0.2047 | 0.4889 | |
| SRP133303 | ARHGAP15 | 55843 | RNAseq | -0.5889 | 0.0277 | |
| SRP159526 | ARHGAP15 | 55843 | RNAseq | -0.6640 | 0.3431 | |
| SRP193095 | ARHGAP15 | 55843 | RNAseq | 0.1822 | 0.5637 | |
| SRP219564 | ARHGAP15 | 55843 | RNAseq | 0.1918 | 0.7665 | |
| TCGA | ARHGAP15 | 55843 | RNAseq | -0.3181 | 0.1314 |
Upregulated datasets: 0; Downregulated datasets: 0.
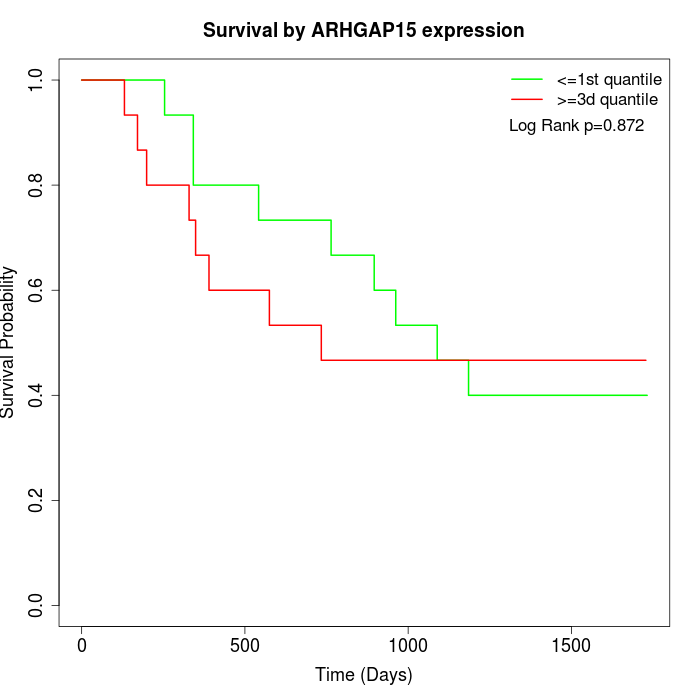
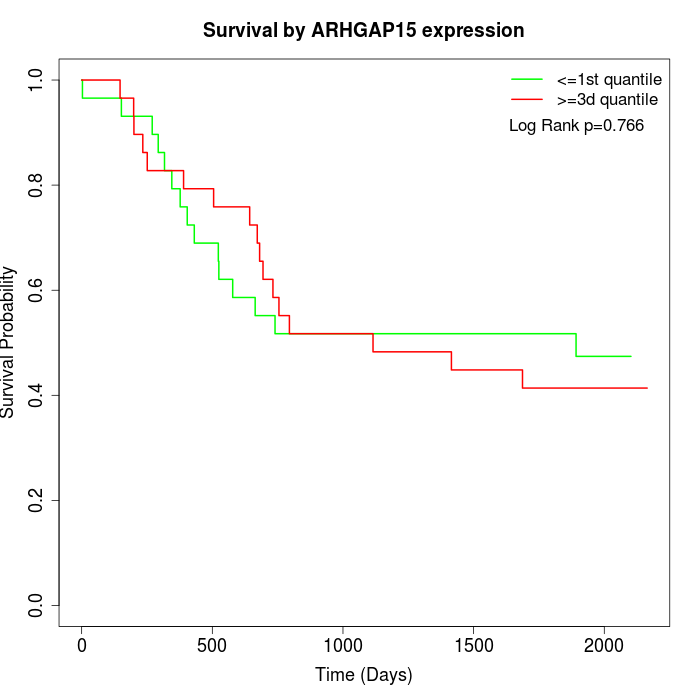
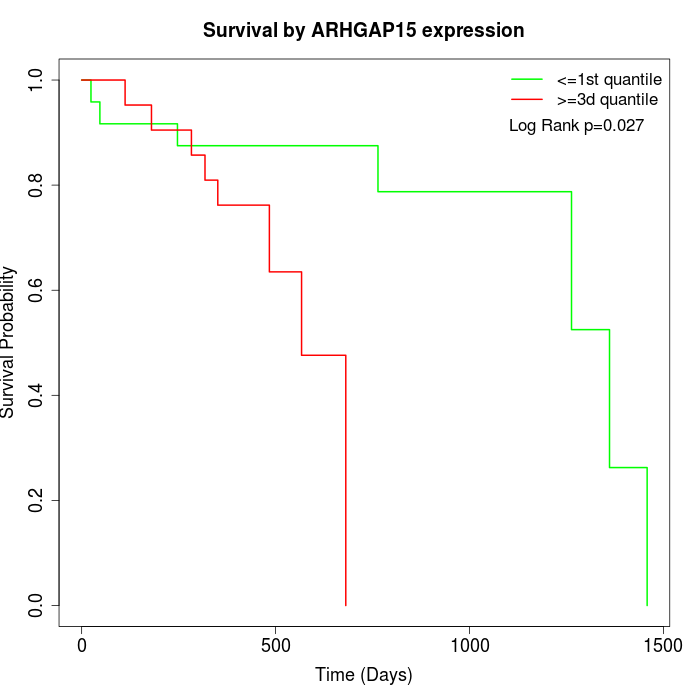
Survival by ARHGAP15 expression:
Note: Click image to view full size file.
Copy number change of ARHGAP15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGAP15 | 55843 | 4 | 4 | 22 | |
| GSE20123 | ARHGAP15 | 55843 | 3 | 4 | 23 | |
| GSE43470 | ARHGAP15 | 55843 | 3 | 1 | 39 | |
| GSE46452 | ARHGAP15 | 55843 | 1 | 4 | 54 | |
| GSE47630 | ARHGAP15 | 55843 | 5 | 3 | 32 | |
| GSE54993 | ARHGAP15 | 55843 | 0 | 5 | 65 | |
| GSE54994 | ARHGAP15 | 55843 | 10 | 2 | 41 | |
| GSE60625 | ARHGAP15 | 55843 | 0 | 3 | 8 | |
| GSE74703 | ARHGAP15 | 55843 | 2 | 1 | 33 | |
| GSE74704 | ARHGAP15 | 55843 | 3 | 2 | 15 | |
| TCGA | ARHGAP15 | 55843 | 22 | 11 | 63 |
Total number of gains: 53; Total number of losses: 40; Total Number of normals: 395.
Somatic mutations of ARHGAP15:
Generating mutation plots.
Highly correlated genes for ARHGAP15:
Showing top 20/344 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGAP15 | GIMAP4 | 0.826404 | 4 | 0 | 4 |
| ARHGAP15 | GRAP2 | 0.813578 | 3 | 0 | 3 |
| ARHGAP15 | GIMAP5 | 0.805699 | 4 | 0 | 4 |
| ARHGAP15 | GIMAP7 | 0.802448 | 4 | 0 | 4 |
| ARHGAP15 | KLRG1 | 0.802302 | 3 | 0 | 3 |
| ARHGAP15 | ARHGEF6 | 0.800263 | 4 | 0 | 4 |
| ARHGAP15 | ANKRD44 | 0.799101 | 3 | 0 | 3 |
| ARHGAP15 | GAPT | 0.797853 | 3 | 0 | 3 |
| ARHGAP15 | GIMAP6 | 0.796802 | 4 | 0 | 4 |
| ARHGAP15 | INPP5D | 0.790879 | 4 | 0 | 4 |
| ARHGAP15 | BIN2 | 0.785418 | 3 | 0 | 3 |
| ARHGAP15 | ABCD2 | 0.784598 | 3 | 0 | 3 |
| ARHGAP15 | RCSD1 | 0.782683 | 4 | 0 | 4 |
| ARHGAP15 | ITK | 0.781955 | 4 | 0 | 4 |
| ARHGAP15 | GIMAP1 | 0.779264 | 4 | 0 | 4 |
| ARHGAP15 | SAMHD1 | 0.775838 | 3 | 0 | 3 |
| ARHGAP15 | TAGAP | 0.774646 | 3 | 0 | 3 |
| ARHGAP15 | TNFSF8 | 0.770387 | 3 | 0 | 3 |
| ARHGAP15 | SRGN | 0.763601 | 3 | 0 | 3 |
| ARHGAP15 | IRF8 | 0.762193 | 4 | 0 | 4 |
For details and further investigation, click here