| Full name: endothelial cell surface expressed chemotaxis and apoptosis regulator | Alias Symbol: ECSM2|ARIA | ||
| Type: protein-coding gene | Cytoband: 5q31.2 | ||
| Entrez ID: 641700 | HGNC ID: HGNC:35454 | Ensembl Gene: ENSG00000249751 | OMIM ID: 615736 |
| Drug and gene relationship at DGIdb | |||
Expression of ECSCR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ECSCR | 641700 | 227780_s_at | -0.5295 | 0.3168 | |
| GSE26886 | ECSCR | 641700 | 227780_s_at | -0.1175 | 0.6178 | |
| GSE45670 | ECSCR | 641700 | 228339_at | -0.8666 | 0.0264 | |
| GSE53622 | ECSCR | 641700 | 90518 | -1.0676 | 0.0000 | |
| GSE53624 | ECSCR | 641700 | 90518 | -0.6089 | 0.0013 | |
| GSE63941 | ECSCR | 641700 | 227780_s_at | 0.1582 | 0.3523 | |
| GSE77861 | ECSCR | 641700 | 227780_s_at | 0.1770 | 0.2989 | |
| GSE97050 | ECSCR | 641700 | A_23_P72651 | -0.0474 | 0.8762 | |
| SRP007169 | ECSCR | 641700 | RNAseq | 0.6491 | 0.2899 | |
| SRP064894 | ECSCR | 641700 | RNAseq | 0.2751 | 0.4989 | |
| SRP133303 | ECSCR | 641700 | RNAseq | -0.9725 | 0.0031 | |
| SRP159526 | ECSCR | 641700 | RNAseq | -0.2866 | 0.5576 | |
| SRP193095 | ECSCR | 641700 | RNAseq | 0.7424 | 0.0046 | |
| SRP219564 | ECSCR | 641700 | RNAseq | -0.2572 | 0.7204 | |
| TCGA | ECSCR | 641700 | RNAseq | -0.5391 | 0.0016 |
Upregulated datasets: 0; Downregulated datasets: 1.
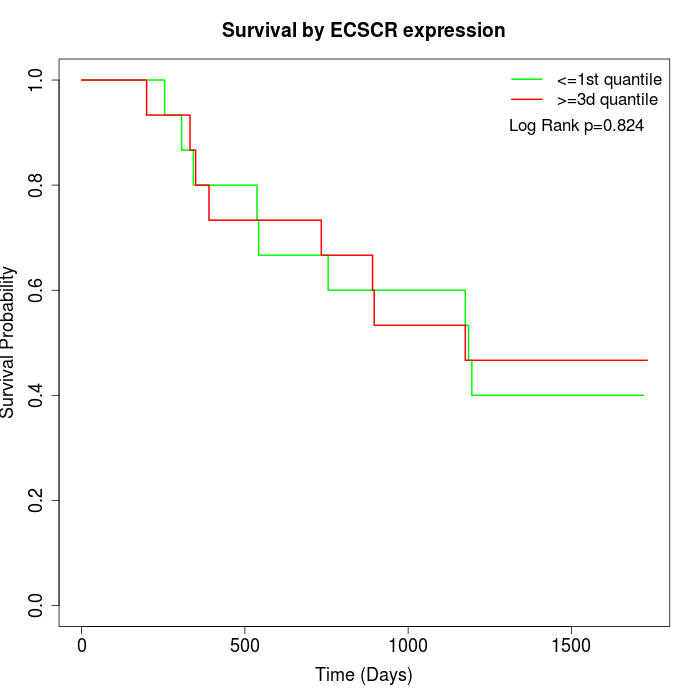
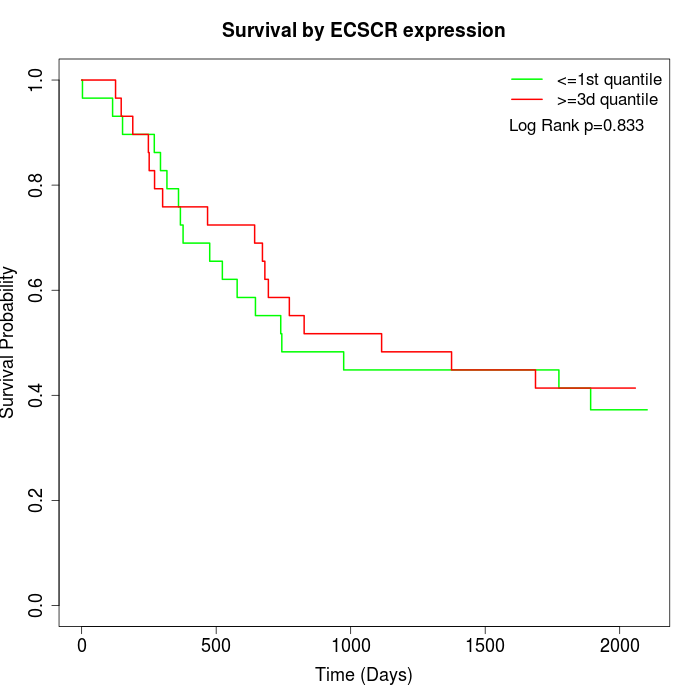
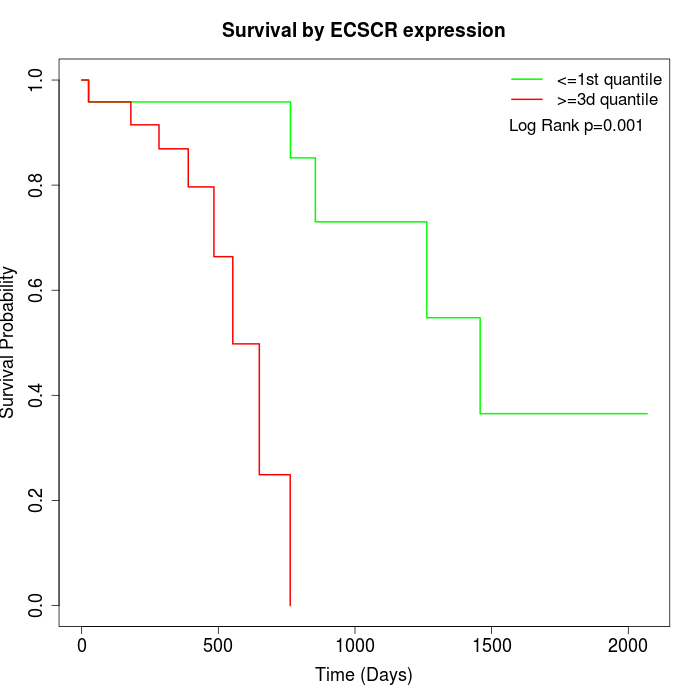
Survival by ECSCR expression:
Note: Click image to view full size file.
Copy number change of ECSCR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ECSCR | 641700 | 3 | 11 | 16 | |
| GSE20123 | ECSCR | 641700 | 4 | 11 | 15 | |
| GSE43470 | ECSCR | 641700 | 4 | 8 | 31 | |
| GSE46452 | ECSCR | 641700 | 0 | 27 | 32 | |
| GSE47630 | ECSCR | 641700 | 0 | 20 | 20 | |
| GSE54993 | ECSCR | 641700 | 9 | 1 | 60 | |
| GSE54994 | ECSCR | 641700 | 2 | 14 | 37 | |
| GSE60625 | ECSCR | 641700 | 0 | 0 | 11 | |
| GSE74703 | ECSCR | 641700 | 3 | 6 | 27 | |
| GSE74704 | ECSCR | 641700 | 3 | 5 | 12 | |
| TCGA | ECSCR | 641700 | 4 | 37 | 55 |
Total number of gains: 32; Total number of losses: 140; Total Number of normals: 316.
Somatic mutations of ECSCR:
Generating mutation plots.
Highly correlated genes for ECSCR:
Showing top 20/576 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ECSCR | PTPRB | 0.872924 | 5 | 0 | 5 |
| ECSCR | GIMAP5 | 0.838712 | 3 | 0 | 3 |
| ECSCR | GNG11 | 0.814884 | 7 | 0 | 7 |
| ECSCR | CD93 | 0.798741 | 8 | 0 | 8 |
| ECSCR | PREX2 | 0.79723 | 4 | 0 | 4 |
| ECSCR | CLEC1A | 0.790963 | 5 | 0 | 5 |
| ECSCR | HGF | 0.783413 | 3 | 0 | 3 |
| ECSCR | TIE1 | 0.782997 | 6 | 0 | 6 |
| ECSCR | ACKR1 | 0.780137 | 6 | 0 | 6 |
| ECSCR | SH3BP5 | 0.779105 | 3 | 0 | 3 |
| ECSCR | EBF3 | 0.775565 | 5 | 0 | 5 |
| ECSCR | CLEC14A | 0.775389 | 7 | 0 | 6 |
| ECSCR | TEK | 0.774279 | 6 | 0 | 5 |
| ECSCR | RRAGA | 0.767784 | 3 | 0 | 3 |
| ECSCR | EPDR1 | 0.767662 | 4 | 0 | 4 |
| ECSCR | SPON1 | 0.762879 | 6 | 0 | 6 |
| ECSCR | C11orf96 | 0.756538 | 3 | 0 | 3 |
| ECSCR | MEOX2 | 0.753172 | 5 | 0 | 5 |
| ECSCR | TNFSF12 | 0.751627 | 3 | 0 | 3 |
| ECSCR | JAK2 | 0.745563 | 3 | 0 | 3 |
For details and further investigation, click here