| Full name: UDP-glucose glycoprotein glucosyltransferase 2 | Alias Symbol: FLJ11485|HUGT2|FLJ10873|MGC150689|MGC87276|MGC117360 | ||
| Type: protein-coding gene | Cytoband: 13q32.1 | ||
| Entrez ID: 55757 | HGNC ID: HGNC:15664 | Ensembl Gene: ENSG00000102595 | OMIM ID: 605898 |
| Drug and gene relationship at DGIdb | |||
Expression of UGGT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | UGGT2 | 55757 | 218801_at | 0.1154 | 0.8155 | |
| GSE20347 | UGGT2 | 55757 | 218801_at | 0.2401 | 0.1430 | |
| GSE23400 | UGGT2 | 55757 | 218801_at | 0.1817 | 0.0000 | |
| GSE26886 | UGGT2 | 55757 | 218801_at | 0.8219 | 0.0005 | |
| GSE29001 | UGGT2 | 55757 | 218801_at | 0.1160 | 0.5458 | |
| GSE38129 | UGGT2 | 55757 | 218801_at | 0.2898 | 0.0955 | |
| GSE45670 | UGGT2 | 55757 | 218801_at | 0.0730 | 0.8169 | |
| GSE53622 | UGGT2 | 55757 | 9022 | 0.4879 | 0.0000 | |
| GSE53624 | UGGT2 | 55757 | 115314 | 0.4786 | 0.0000 | |
| GSE63941 | UGGT2 | 55757 | 235749_at | 0.4850 | 0.5236 | |
| GSE77861 | UGGT2 | 55757 | 218801_at | 0.2208 | 0.1840 | |
| GSE97050 | UGGT2 | 55757 | A_24_P44341 | 0.3228 | 0.2199 | |
| SRP007169 | UGGT2 | 55757 | RNAseq | 0.5339 | 0.1112 | |
| SRP008496 | UGGT2 | 55757 | RNAseq | 1.1997 | 0.0000 | |
| SRP064894 | UGGT2 | 55757 | RNAseq | 0.7147 | 0.0001 | |
| SRP133303 | UGGT2 | 55757 | RNAseq | 0.4657 | 0.0131 | |
| SRP159526 | UGGT2 | 55757 | RNAseq | 0.4233 | 0.0774 | |
| SRP193095 | UGGT2 | 55757 | RNAseq | 0.1881 | 0.2076 | |
| SRP219564 | UGGT2 | 55757 | RNAseq | 0.5323 | 0.1083 | |
| TCGA | UGGT2 | 55757 | RNAseq | -0.1984 | 0.0036 |
Upregulated datasets: 1; Downregulated datasets: 0.
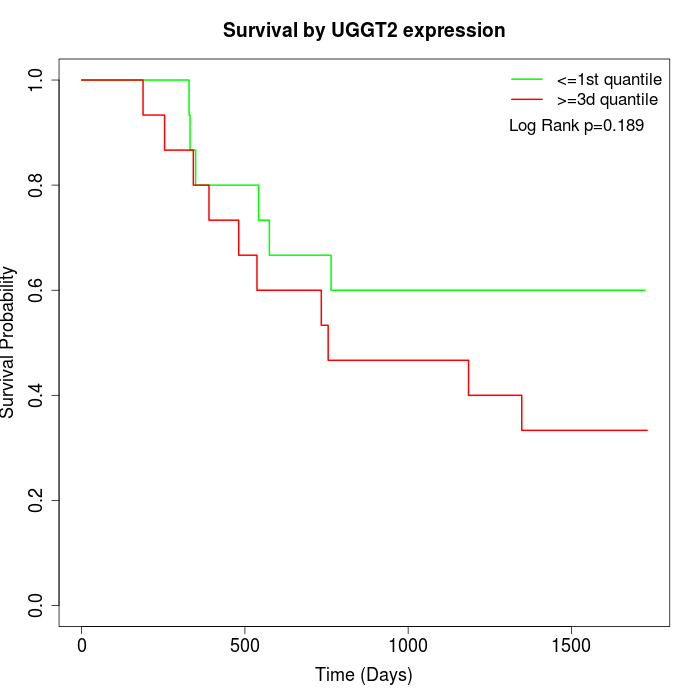
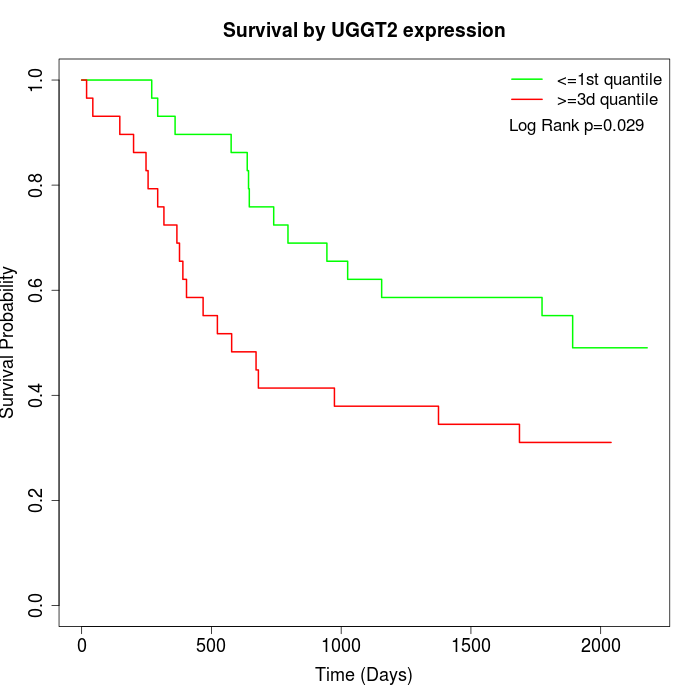
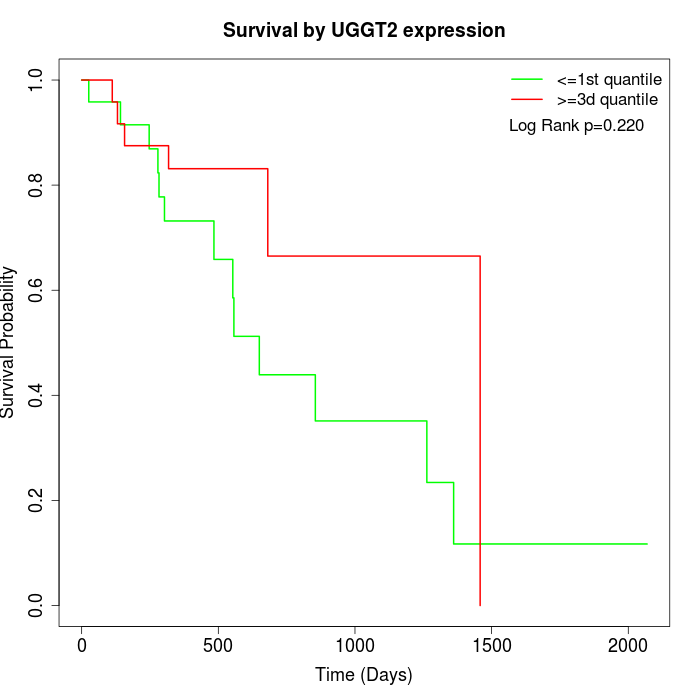
Survival by UGGT2 expression:
Note: Click image to view full size file.
Copy number change of UGGT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | UGGT2 | 55757 | 4 | 9 | 17 | |
| GSE20123 | UGGT2 | 55757 | 4 | 8 | 18 | |
| GSE43470 | UGGT2 | 55757 | 4 | 12 | 27 | |
| GSE46452 | UGGT2 | 55757 | 0 | 32 | 27 | |
| GSE47630 | UGGT2 | 55757 | 3 | 27 | 10 | |
| GSE54993 | UGGT2 | 55757 | 11 | 3 | 56 | |
| GSE54994 | UGGT2 | 55757 | 3 | 13 | 37 | |
| GSE60625 | UGGT2 | 55757 | 0 | 3 | 8 | |
| GSE74703 | UGGT2 | 55757 | 3 | 10 | 23 | |
| GSE74704 | UGGT2 | 55757 | 2 | 8 | 10 | |
| TCGA | UGGT2 | 55757 | 13 | 34 | 49 |
Total number of gains: 47; Total number of losses: 159; Total Number of normals: 282.
Somatic mutations of UGGT2:
Generating mutation plots.
Highly correlated genes for UGGT2:
Showing top 20/995 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| UGGT2 | SLC39A1 | 0.789872 | 3 | 0 | 3 |
| UGGT2 | CHERP | 0.757355 | 3 | 0 | 3 |
| UGGT2 | ZNF703 | 0.7518 | 3 | 0 | 3 |
| UGGT2 | B3GALT6 | 0.741276 | 3 | 0 | 3 |
| UGGT2 | PTGS2 | 0.74001 | 3 | 0 | 3 |
| UGGT2 | SORBS3 | 0.733705 | 3 | 0 | 3 |
| UGGT2 | TMTC4 | 0.732058 | 4 | 0 | 3 |
| UGGT2 | CEP70 | 0.731703 | 3 | 0 | 3 |
| UGGT2 | DCAF7 | 0.728122 | 3 | 0 | 3 |
| UGGT2 | DERL2 | 0.727798 | 3 | 0 | 3 |
| UGGT2 | METTL23 | 0.726415 | 4 | 0 | 4 |
| UGGT2 | KCNE3 | 0.722552 | 3 | 0 | 3 |
| UGGT2 | DHRSX | 0.722364 | 4 | 0 | 3 |
| UGGT2 | CAPN12 | 0.718824 | 4 | 0 | 3 |
| UGGT2 | ZFYVE27 | 0.715316 | 4 | 0 | 3 |
| UGGT2 | ZNF121 | 0.71514 | 3 | 0 | 3 |
| UGGT2 | UROS | 0.715093 | 3 | 0 | 3 |
| UGGT2 | ZNF704 | 0.71417 | 3 | 0 | 3 |
| UGGT2 | TMTC1 | 0.710924 | 3 | 0 | 3 |
| UGGT2 | WDR81 | 0.710653 | 4 | 0 | 3 |
For details and further investigation, click here