| Full name: delta 4-desaturase, sphingolipid 1 | Alias Symbol: MLD|Des-1|DES1|FADS7|DEGS-1 | ||
| Type: protein-coding gene | Cytoband: 1q42.11 | ||
| Entrez ID: 8560 | HGNC ID: HGNC:13709 | Ensembl Gene: ENSG00000143753 | OMIM ID: 615843 |
| Drug and gene relationship at DGIdb | |||
Expression of DEGS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DEGS1 | 8560 | 209250_at | 0.3943 | 0.6992 | |
| GSE20347 | DEGS1 | 8560 | 209250_at | 0.3701 | 0.0696 | |
| GSE23400 | DEGS1 | 8560 | 209250_at | 0.5648 | 0.0000 | |
| GSE26886 | DEGS1 | 8560 | 209250_at | 1.6132 | 0.0000 | |
| GSE29001 | DEGS1 | 8560 | 209250_at | 0.4949 | 0.2363 | |
| GSE38129 | DEGS1 | 8560 | 209250_at | 0.1809 | 0.2628 | |
| GSE45670 | DEGS1 | 8560 | 209250_at | 0.0889 | 0.8106 | |
| GSE53622 | DEGS1 | 8560 | 58103 | 0.1710 | 0.0302 | |
| GSE53624 | DEGS1 | 8560 | 58103 | 0.3844 | 0.0000 | |
| GSE63941 | DEGS1 | 8560 | 209250_at | -2.2285 | 0.0057 | |
| GSE77861 | DEGS1 | 8560 | 209250_at | 0.5291 | 0.1447 | |
| GSE97050 | DEGS1 | 8560 | A_33_P3290179 | -0.0521 | 0.8643 | |
| SRP007169 | DEGS1 | 8560 | RNAseq | -0.0573 | 0.8878 | |
| SRP008496 | DEGS1 | 8560 | RNAseq | 0.2896 | 0.2852 | |
| SRP064894 | DEGS1 | 8560 | RNAseq | 0.1806 | 0.2944 | |
| SRP133303 | DEGS1 | 8560 | RNAseq | 0.1711 | 0.3887 | |
| SRP159526 | DEGS1 | 8560 | RNAseq | 0.2849 | 0.4764 | |
| SRP193095 | DEGS1 | 8560 | RNAseq | 0.1973 | 0.2146 | |
| SRP219564 | DEGS1 | 8560 | RNAseq | -0.1551 | 0.7091 | |
| TCGA | DEGS1 | 8560 | RNAseq | 0.1023 | 0.0870 |
Upregulated datasets: 1; Downregulated datasets: 1.
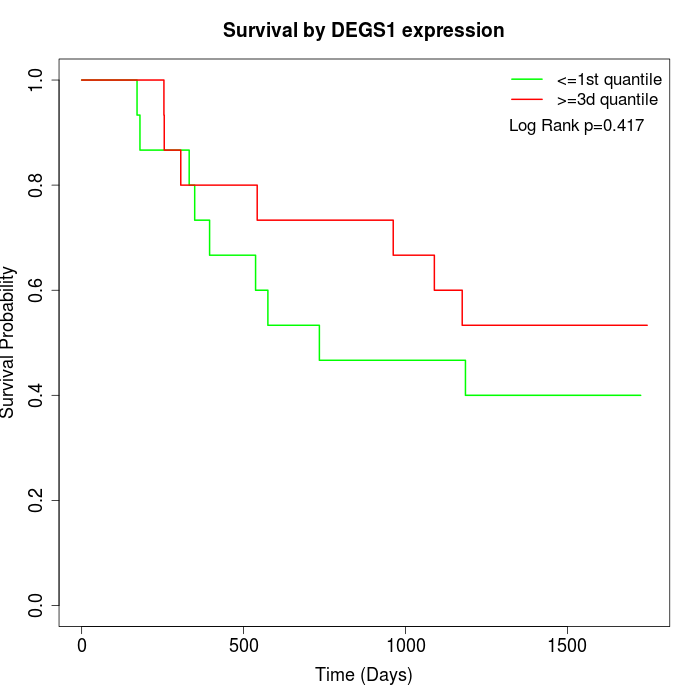
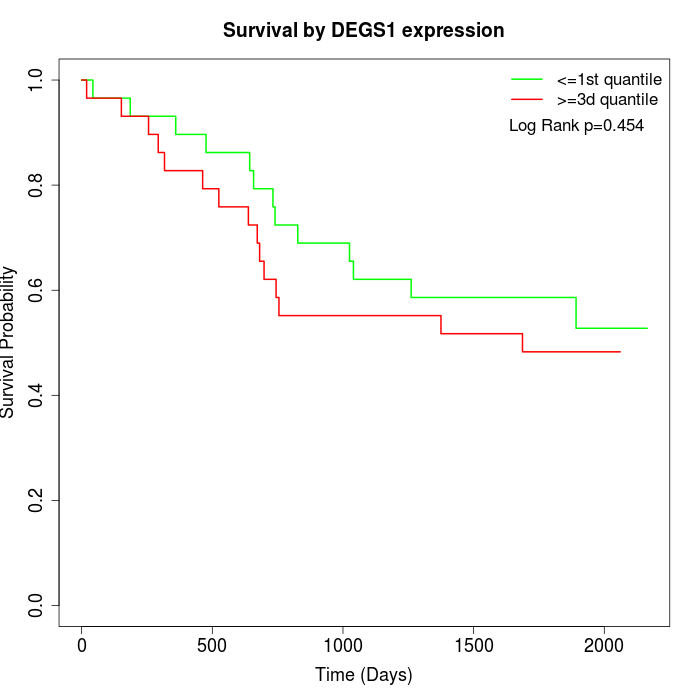
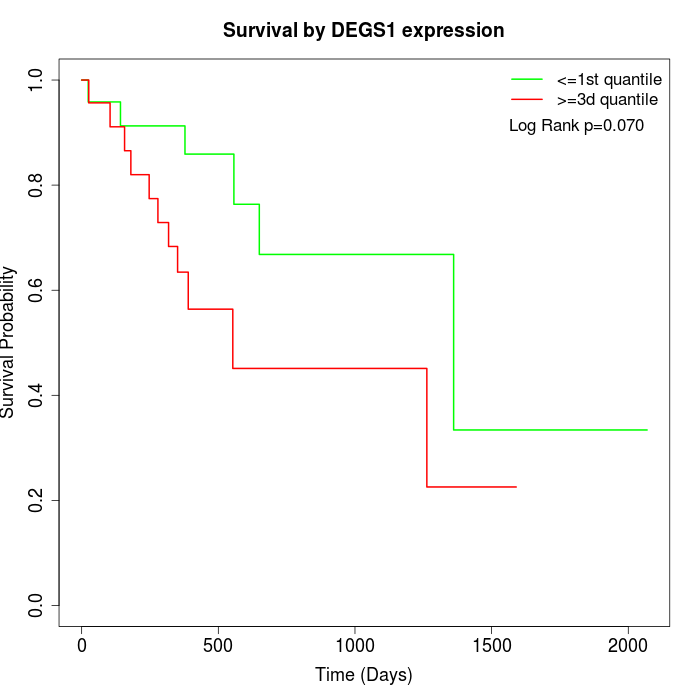
Survival by DEGS1 expression:
Note: Click image to view full size file.
Copy number change of DEGS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DEGS1 | 8560 | 10 | 0 | 20 | |
| GSE20123 | DEGS1 | 8560 | 10 | 0 | 20 | |
| GSE43470 | DEGS1 | 8560 | 7 | 0 | 36 | |
| GSE46452 | DEGS1 | 8560 | 3 | 1 | 55 | |
| GSE47630 | DEGS1 | 8560 | 15 | 0 | 25 | |
| GSE54993 | DEGS1 | 8560 | 0 | 6 | 64 | |
| GSE54994 | DEGS1 | 8560 | 16 | 0 | 37 | |
| GSE60625 | DEGS1 | 8560 | 0 | 0 | 11 | |
| GSE74703 | DEGS1 | 8560 | 7 | 0 | 29 | |
| GSE74704 | DEGS1 | 8560 | 4 | 0 | 16 | |
| TCGA | DEGS1 | 8560 | 45 | 4 | 47 |
Total number of gains: 117; Total number of losses: 11; Total Number of normals: 360.
Somatic mutations of DEGS1:
Generating mutation plots.
Highly correlated genes for DEGS1:
Showing top 20/784 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DEGS1 | FOXN3 | 0.827314 | 3 | 0 | 3 |
| DEGS1 | ZCCHC24 | 0.798363 | 3 | 0 | 3 |
| DEGS1 | EEF1D | 0.785038 | 3 | 0 | 3 |
| DEGS1 | DPP4 | 0.780986 | 3 | 0 | 3 |
| DEGS1 | FAM20C | 0.779952 | 4 | 0 | 4 |
| DEGS1 | LIX1L | 0.770932 | 3 | 0 | 3 |
| DEGS1 | PKIG | 0.764854 | 4 | 0 | 4 |
| DEGS1 | HABP4 | 0.750987 | 3 | 0 | 3 |
| DEGS1 | CRISPLD2 | 0.749314 | 4 | 0 | 4 |
| DEGS1 | CWC22 | 0.746256 | 3 | 0 | 3 |
| DEGS1 | SH2B3 | 0.744301 | 3 | 0 | 3 |
| DEGS1 | NBPF20 | 0.741221 | 3 | 0 | 3 |
| DEGS1 | SEC22A | 0.738547 | 3 | 0 | 3 |
| DEGS1 | AP3B2 | 0.737594 | 3 | 0 | 3 |
| DEGS1 | NKAP | 0.734773 | 3 | 0 | 3 |
| DEGS1 | ANKS3 | 0.734381 | 3 | 0 | 3 |
| DEGS1 | MAPK9 | 0.729221 | 3 | 0 | 3 |
| DEGS1 | TMEM176A | 0.727671 | 3 | 0 | 3 |
| DEGS1 | GPATCH1 | 0.726565 | 3 | 0 | 3 |
| DEGS1 | C11orf96 | 0.726272 | 3 | 0 | 3 |
For details and further investigation, click here