| Full name: GTPase, IMAP family member 5 | Alias Symbol: HIMAP3|IAN5 | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 55340 | HGNC ID: HGNC:18005 | Ensembl Gene: ENSG00000196329 | OMIM ID: 608086 |
| Drug and gene relationship at DGIdb | |||
Expression of GIMAP5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | GIMAP5 | 55340 | 72531 | -0.9610 | 0.0000 | |
| GSE53624 | GIMAP5 | 55340 | 72531 | -0.8292 | 0.0000 | |
| GSE97050 | GIMAP5 | 55340 | A_23_P42588 | 0.0333 | 0.9444 | |
| SRP007169 | GIMAP5 | 55340 | RNAseq | 0.5599 | 0.5517 | |
| SRP008496 | GIMAP5 | 55340 | RNAseq | 0.4236 | 0.3625 | |
| SRP064894 | GIMAP5 | 55340 | RNAseq | 0.1570 | 0.7165 | |
| SRP133303 | GIMAP5 | 55340 | RNAseq | -0.5117 | 0.1288 | |
| SRP159526 | GIMAP5 | 55340 | RNAseq | -0.7762 | 0.1005 | |
| SRP193095 | GIMAP5 | 55340 | RNAseq | -0.2061 | 0.4556 | |
| SRP219564 | GIMAP5 | 55340 | RNAseq | 0.1559 | 0.7651 | |
| TCGA | GIMAP5 | 55340 | RNAseq | -0.5477 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 0.
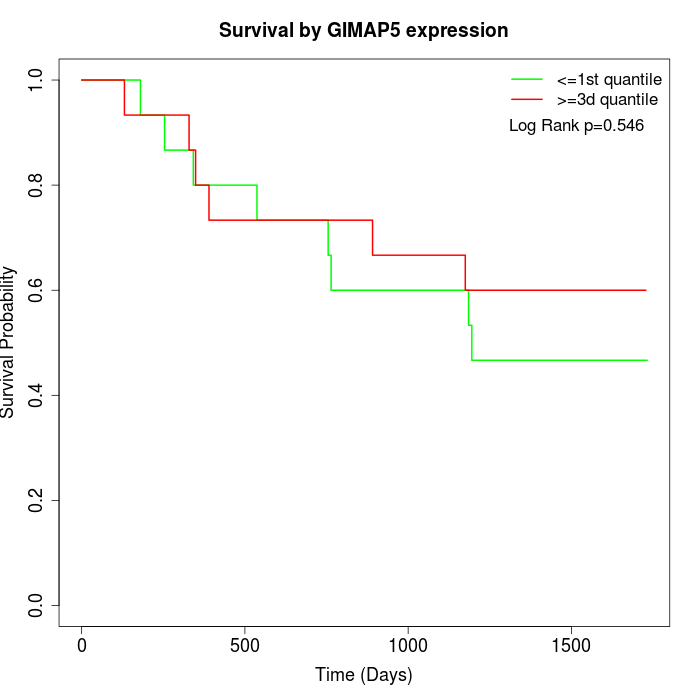
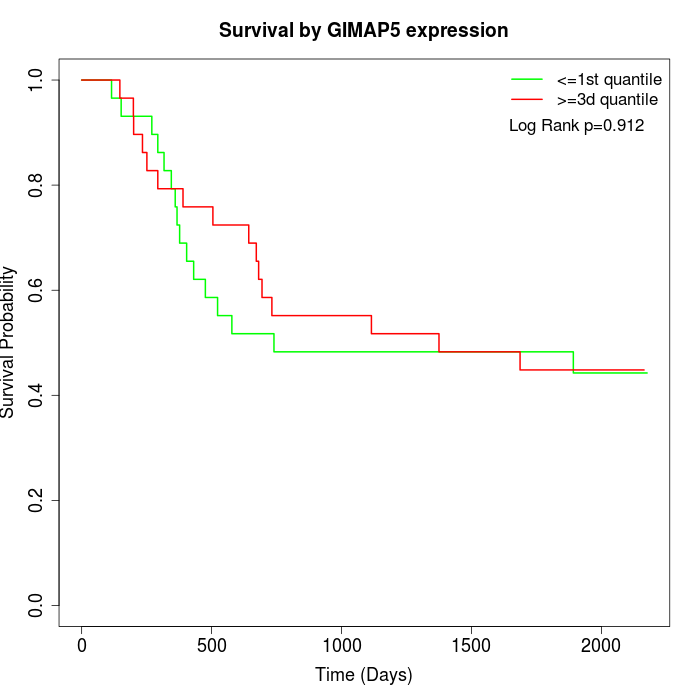
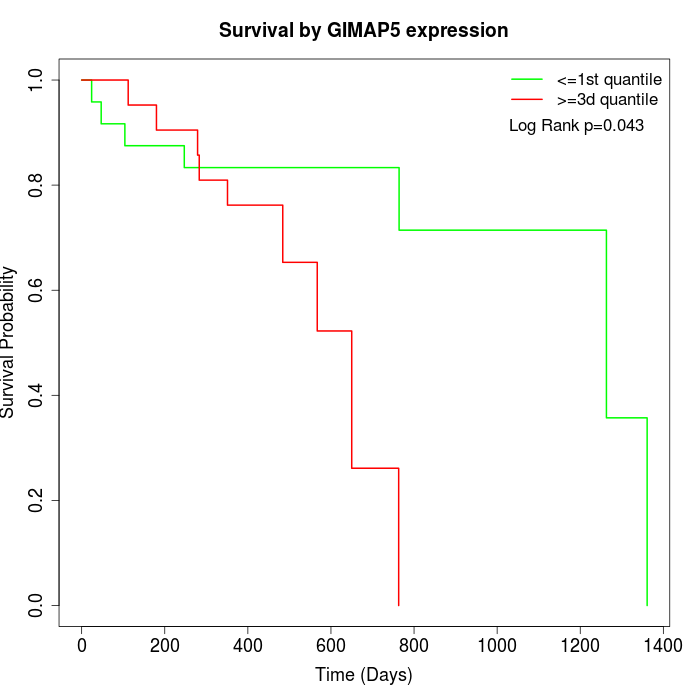
Survival by GIMAP5 expression:
Note: Click image to view full size file.
Copy number change of GIMAP5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GIMAP5 | 55340 | 2 | 4 | 24 | |
| GSE20123 | GIMAP5 | 55340 | 2 | 4 | 24 | |
| GSE43470 | GIMAP5 | 55340 | 2 | 5 | 36 | |
| GSE46452 | GIMAP5 | 55340 | 7 | 2 | 50 | |
| GSE47630 | GIMAP5 | 55340 | 5 | 9 | 26 | |
| GSE54993 | GIMAP5 | 55340 | 3 | 3 | 64 | |
| GSE54994 | GIMAP5 | 55340 | 5 | 8 | 40 | |
| GSE60625 | GIMAP5 | 55340 | 0 | 0 | 11 | |
| GSE74703 | GIMAP5 | 55340 | 2 | 4 | 30 | |
| GSE74704 | GIMAP5 | 55340 | 1 | 4 | 15 | |
| TCGA | GIMAP5 | 55340 | 25 | 26 | 45 |
Total number of gains: 54; Total number of losses: 69; Total Number of normals: 365.
Somatic mutations of GIMAP5:
Generating mutation plots.
Highly correlated genes for GIMAP5:
Showing top 20/447 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GIMAP5 | GIMAP7 | 0.870496 | 4 | 0 | 4 |
| GIMAP5 | GIMAP4 | 0.855026 | 4 | 0 | 4 |
| GIMAP5 | CD226 | 0.845539 | 3 | 0 | 3 |
| GIMAP5 | ECSCR | 0.838712 | 3 | 0 | 3 |
| GIMAP5 | RGS18 | 0.834368 | 3 | 0 | 3 |
| GIMAP5 | LILRB5 | 0.829928 | 3 | 0 | 3 |
| GIMAP5 | MMRN1 | 0.82991 | 3 | 0 | 3 |
| GIMAP5 | MPEG1 | 0.829605 | 3 | 0 | 3 |
| GIMAP5 | ITK | 0.821662 | 4 | 0 | 4 |
| GIMAP5 | GAB3 | 0.821518 | 4 | 0 | 4 |
| GIMAP5 | TNFSF8 | 0.82112 | 3 | 0 | 3 |
| GIMAP5 | C16orf54 | 0.82077 | 3 | 0 | 3 |
| GIMAP5 | GIMAP1 | 0.818436 | 4 | 0 | 3 |
| GIMAP5 | GIMAP8 | 0.818071 | 4 | 0 | 4 |
| GIMAP5 | PTPRB | 0.817838 | 3 | 0 | 3 |
| GIMAP5 | IRF8 | 0.81592 | 4 | 0 | 4 |
| GIMAP5 | NCKAP1L | 0.811269 | 4 | 0 | 4 |
| GIMAP5 | CPVL | 0.808603 | 3 | 0 | 3 |
| GIMAP5 | TFPI | 0.80839 | 3 | 0 | 3 |
| GIMAP5 | HGF | 0.807194 | 3 | 0 | 3 |
For details and further investigation, click here