| Full name: calsyntenin 2 | Alias Symbol: CSTN2|CS2|FLJ39113|CDHR13 | ||
| Type: protein-coding gene | Cytoband: 3q23 | ||
| Entrez ID: 64084 | HGNC ID: HGNC:17448 | Ensembl Gene: ENSG00000158258 | OMIM ID: 611323 |
| Drug and gene relationship at DGIdb | |||
Expression of CLSTN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLSTN2 | 64084 | 229580_at | -0.5944 | 0.6149 | |
| GSE20347 | CLSTN2 | 64084 | 219414_at | 0.1912 | 0.0417 | |
| GSE23400 | CLSTN2 | 64084 | 219414_at | -0.0483 | 0.3316 | |
| GSE26886 | CLSTN2 | 64084 | 229580_at | 0.8669 | 0.0194 | |
| GSE29001 | CLSTN2 | 64084 | 219414_at | 0.0445 | 0.8378 | |
| GSE38129 | CLSTN2 | 64084 | 219414_at | 0.0977 | 0.4986 | |
| GSE45670 | CLSTN2 | 64084 | 229580_at | -1.1355 | 0.0006 | |
| GSE53622 | CLSTN2 | 64084 | 7171 | -0.3360 | 0.1249 | |
| GSE53624 | CLSTN2 | 64084 | 7171 | 0.1362 | 0.5352 | |
| GSE63941 | CLSTN2 | 64084 | 229580_at | -0.0446 | 0.9709 | |
| GSE77861 | CLSTN2 | 64084 | 219414_at | 0.1210 | 0.4459 | |
| GSE97050 | CLSTN2 | 64084 | A_23_P212608 | 0.3107 | 0.6253 | |
| SRP007169 | CLSTN2 | 64084 | RNAseq | 3.7704 | 0.0000 | |
| SRP008496 | CLSTN2 | 64084 | RNAseq | 2.7401 | 0.0000 | |
| SRP064894 | CLSTN2 | 64084 | RNAseq | -0.3789 | 0.3195 | |
| SRP133303 | CLSTN2 | 64084 | RNAseq | -0.0157 | 0.9574 | |
| SRP159526 | CLSTN2 | 64084 | RNAseq | -0.3575 | 0.5004 | |
| SRP193095 | CLSTN2 | 64084 | RNAseq | 2.0718 | 0.0000 | |
| SRP219564 | CLSTN2 | 64084 | RNAseq | -0.6811 | 0.2714 | |
| TCGA | CLSTN2 | 64084 | RNAseq | 0.1007 | 0.6758 |
Upregulated datasets: 3; Downregulated datasets: 1.
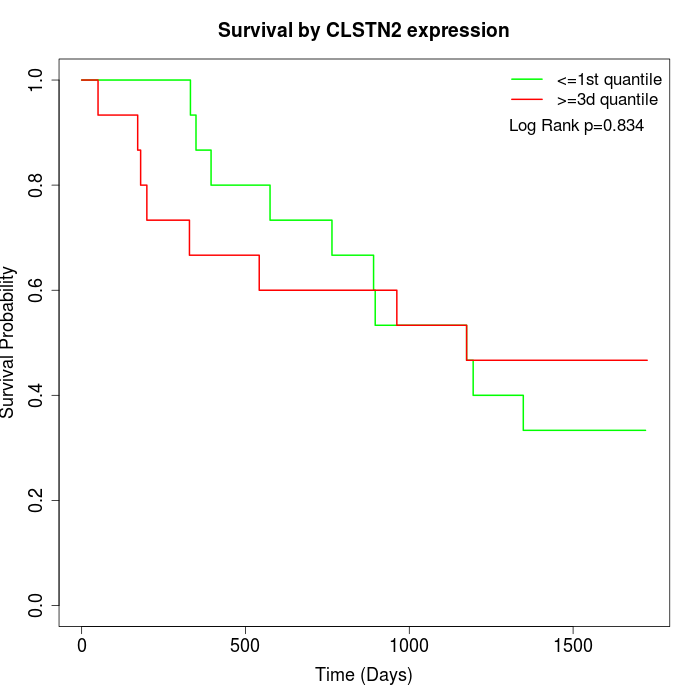
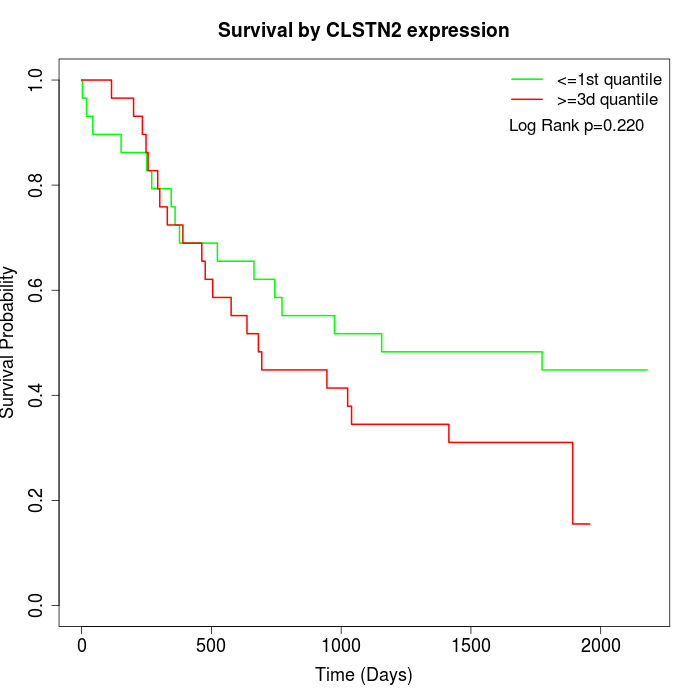
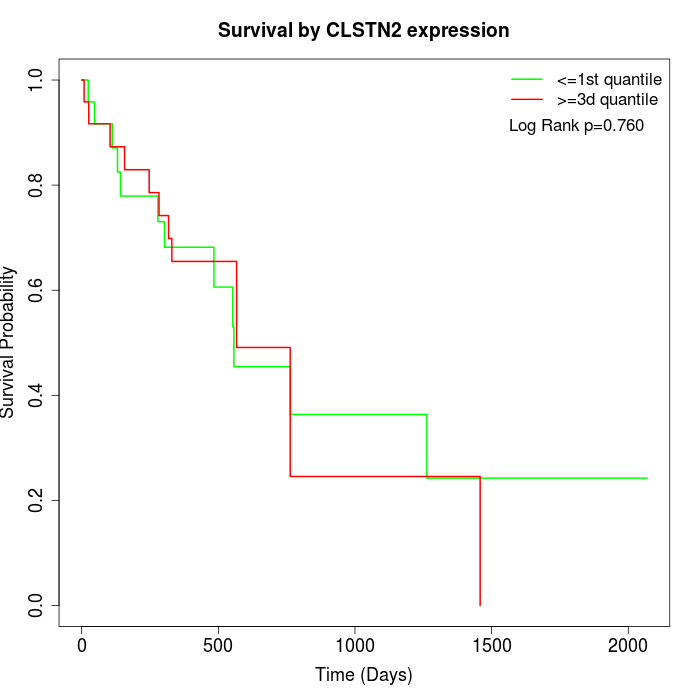
Survival by CLSTN2 expression:
Note: Click image to view full size file.
Copy number change of CLSTN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLSTN2 | 64084 | 19 | 0 | 11 | |
| GSE20123 | CLSTN2 | 64084 | 19 | 0 | 11 | |
| GSE43470 | CLSTN2 | 64084 | 22 | 0 | 21 | |
| GSE46452 | CLSTN2 | 64084 | 15 | 4 | 40 | |
| GSE47630 | CLSTN2 | 64084 | 19 | 3 | 18 | |
| GSE54993 | CLSTN2 | 64084 | 1 | 8 | 61 | |
| GSE54994 | CLSTN2 | 64084 | 34 | 2 | 17 | |
| GSE60625 | CLSTN2 | 64084 | 0 | 6 | 5 | |
| GSE74703 | CLSTN2 | 64084 | 19 | 0 | 17 | |
| GSE74704 | CLSTN2 | 64084 | 13 | 0 | 7 | |
| TCGA | CLSTN2 | 64084 | 62 | 6 | 28 |
Total number of gains: 223; Total number of losses: 29; Total Number of normals: 236.
Somatic mutations of CLSTN2:
Generating mutation plots.
Highly correlated genes for CLSTN2:
Showing top 20/582 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLSTN2 | S1PR1 | 0.754963 | 3 | 0 | 3 |
| CLSTN2 | MLLT3 | 0.735885 | 3 | 0 | 3 |
| CLSTN2 | RABAC1 | 0.735782 | 3 | 0 | 3 |
| CLSTN2 | ZNF414 | 0.730535 | 3 | 0 | 3 |
| CLSTN2 | TP53I13 | 0.720202 | 3 | 0 | 3 |
| CLSTN2 | RAB39B | 0.719487 | 3 | 0 | 3 |
| CLSTN2 | SPRY1 | 0.715547 | 3 | 0 | 3 |
| CLSTN2 | MSI1 | 0.713051 | 3 | 0 | 3 |
| CLSTN2 | MAN1A2 | 0.712446 | 3 | 0 | 3 |
| CLSTN2 | GGT5 | 0.703813 | 4 | 0 | 4 |
| CLSTN2 | VAMP1 | 0.703398 | 3 | 0 | 3 |
| CLSTN2 | KCNE3 | 0.70181 | 3 | 0 | 3 |
| CLSTN2 | FABP4 | 0.698073 | 3 | 0 | 3 |
| CLSTN2 | INPP5E | 0.695929 | 3 | 0 | 3 |
| CLSTN2 | ZBTB16 | 0.690857 | 3 | 0 | 3 |
| CLSTN2 | DSEL | 0.690058 | 5 | 0 | 5 |
| CLSTN2 | USP42 | 0.685665 | 3 | 0 | 3 |
| CLSTN2 | TCTEX1D2 | 0.68507 | 4 | 0 | 3 |
| CLSTN2 | NENF | 0.683128 | 4 | 0 | 4 |
| CLSTN2 | CD93 | 0.682416 | 5 | 0 | 5 |
For details and further investigation, click here