| Full name: sphingosine-1-phosphate receptor 1 | Alias Symbol: edg-1|D1S3362|CD363 | ||
| Type: protein-coding gene | Cytoband: 1p21.2 | ||
| Entrez ID: 1901 | HGNC ID: HGNC:3165 | Ensembl Gene: ENSG00000170989 | OMIM ID: 601974 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
S1PR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04071 | Sphingolipid signaling pathway |
Expression of S1PR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | S1PR1 | 1901 | 204642_at | -0.7747 | 0.1241 | |
| GSE20347 | S1PR1 | 1901 | 204642_at | -0.1861 | 0.0590 | |
| GSE23400 | S1PR1 | 1901 | 204642_at | -0.1189 | 0.0027 | |
| GSE26886 | S1PR1 | 1901 | 204642_at | -0.0482 | 0.7460 | |
| GSE29001 | S1PR1 | 1901 | 204642_at | -0.1934 | 0.2611 | |
| GSE38129 | S1PR1 | 1901 | 204642_at | -0.6036 | 0.0040 | |
| GSE45670 | S1PR1 | 1901 | 204642_at | -0.9884 | 0.0002 | |
| GSE53622 | S1PR1 | 1901 | 21983 | -1.4680 | 0.0000 | |
| GSE53624 | S1PR1 | 1901 | 21983 | -1.0509 | 0.0000 | |
| GSE63941 | S1PR1 | 1901 | 204642_at | -2.1890 | 0.0003 | |
| GSE77861 | S1PR1 | 1901 | 204642_at | 0.0829 | 0.3722 | |
| GSE97050 | S1PR1 | 1901 | A_23_P404481 | 0.3052 | 0.3792 | |
| SRP007169 | S1PR1 | 1901 | RNAseq | 0.4435 | 0.3619 | |
| SRP008496 | S1PR1 | 1901 | RNAseq | 0.8915 | 0.0310 | |
| SRP064894 | S1PR1 | 1901 | RNAseq | 0.2314 | 0.5860 | |
| SRP133303 | S1PR1 | 1901 | RNAseq | -0.8893 | 0.0080 | |
| SRP159526 | S1PR1 | 1901 | RNAseq | -0.2602 | 0.6241 | |
| SRP193095 | S1PR1 | 1901 | RNAseq | 0.7330 | 0.0076 | |
| SRP219564 | S1PR1 | 1901 | RNAseq | -0.2779 | 0.6960 | |
| TCGA | S1PR1 | 1901 | RNAseq | -0.7597 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 3.
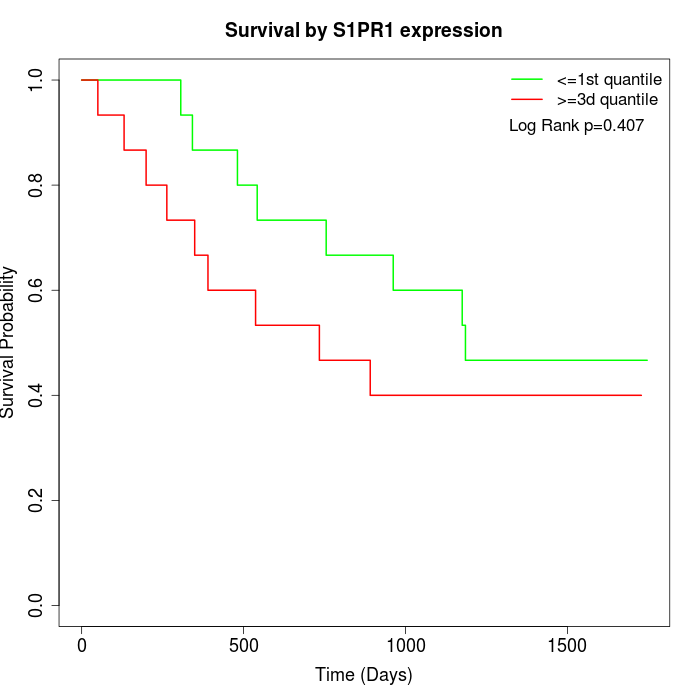
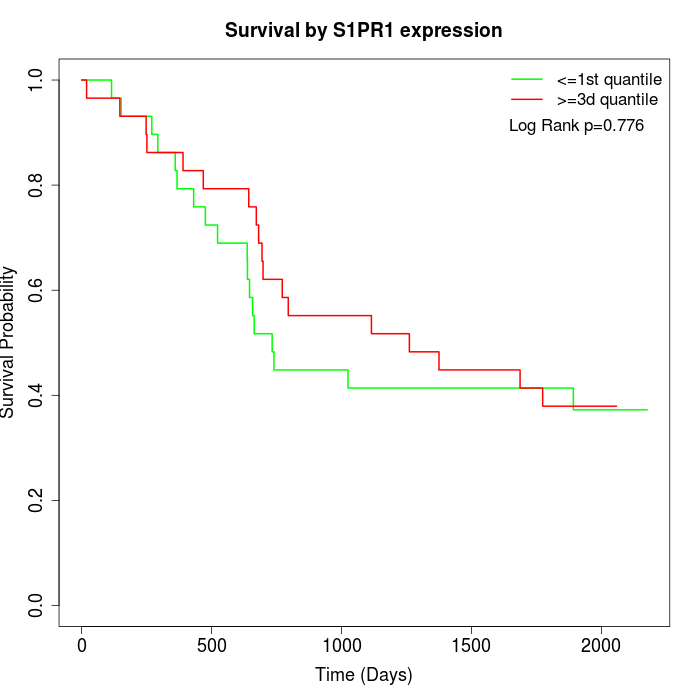
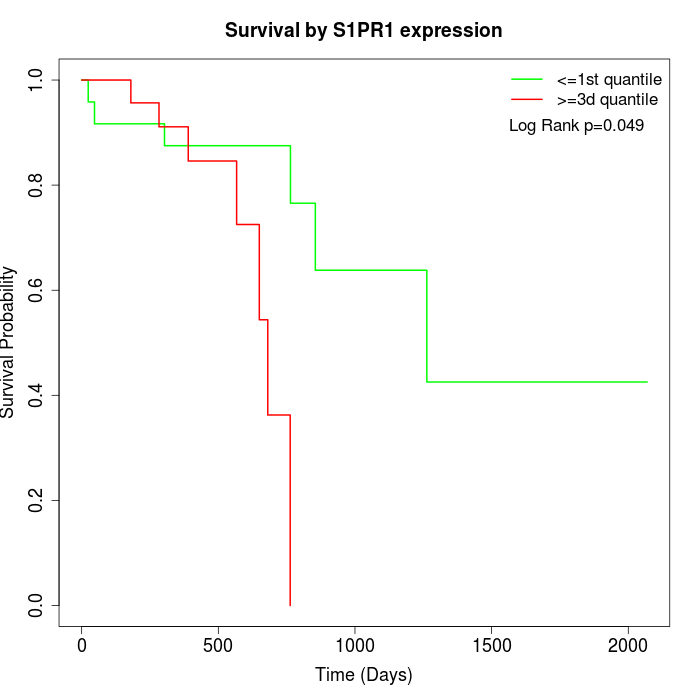
Survival by S1PR1 expression:
Note: Click image to view full size file.
Copy number change of S1PR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | S1PR1 | 1901 | 0 | 10 | 20 | |
| GSE20123 | S1PR1 | 1901 | 0 | 10 | 20 | |
| GSE43470 | S1PR1 | 1901 | 2 | 6 | 35 | |
| GSE46452 | S1PR1 | 1901 | 1 | 1 | 57 | |
| GSE47630 | S1PR1 | 1901 | 9 | 5 | 26 | |
| GSE54993 | S1PR1 | 1901 | 0 | 1 | 69 | |
| GSE54994 | S1PR1 | 1901 | 7 | 3 | 43 | |
| GSE60625 | S1PR1 | 1901 | 0 | 0 | 11 | |
| GSE74703 | S1PR1 | 1901 | 1 | 5 | 30 | |
| GSE74704 | S1PR1 | 1901 | 0 | 6 | 14 | |
| TCGA | S1PR1 | 1901 | 8 | 25 | 63 |
Total number of gains: 28; Total number of losses: 72; Total Number of normals: 388.
Somatic mutations of S1PR1:
Generating mutation plots.
Highly correlated genes for S1PR1:
Showing top 20/1112 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| S1PR1 | SNX25 | 0.840008 | 3 | 0 | 3 |
| S1PR1 | C11orf96 | 0.831616 | 4 | 0 | 4 |
| S1PR1 | CLEC3B | 0.829971 | 3 | 0 | 3 |
| S1PR1 | ZNF583 | 0.829447 | 3 | 0 | 3 |
| S1PR1 | C11orf68 | 0.817623 | 3 | 0 | 3 |
| S1PR1 | KDM5D | 0.813898 | 3 | 0 | 3 |
| S1PR1 | GLIS3 | 0.801394 | 3 | 0 | 3 |
| S1PR1 | TBX5-AS1 | 0.782909 | 3 | 0 | 3 |
| S1PR1 | MAGI2-AS3 | 0.780706 | 6 | 0 | 6 |
| S1PR1 | GFRA1 | 0.77465 | 7 | 0 | 7 |
| S1PR1 | CYP21A2 | 0.773548 | 3 | 0 | 3 |
| S1PR1 | LINC01082 | 0.772226 | 5 | 0 | 5 |
| S1PR1 | COL14A1 | 0.770851 | 10 | 0 | 10 |
| S1PR1 | GIMAP5 | 0.770009 | 3 | 0 | 3 |
| S1PR1 | LINC01279 | 0.764915 | 4 | 0 | 4 |
| S1PR1 | HDHD2 | 0.763889 | 3 | 0 | 3 |
| S1PR1 | ESYT2 | 0.763246 | 3 | 0 | 3 |
| S1PR1 | CYP4V2 | 0.76218 | 4 | 0 | 4 |
| S1PR1 | VEGFC | 0.760293 | 3 | 0 | 3 |
| S1PR1 | DCHS1 | 0.760081 | 6 | 0 | 6 |
For details and further investigation, click here