| Full name: collectin subfamily member 10 | Alias Symbol: CL-L1 | ||
| Type: protein-coding gene | Cytoband: 8q24.12 | ||
| Entrez ID: 10584 | HGNC ID: HGNC:2220 | Ensembl Gene: ENSG00000184374 | OMIM ID: 607620 |
| Drug and gene relationship at DGIdb | |||
Expression of COLEC10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | COLEC10 | 10584 | 207420_at | 0.0115 | 0.9739 | |
| GSE20347 | COLEC10 | 10584 | 207420_at | 0.1534 | 0.1110 | |
| GSE23400 | COLEC10 | 10584 | 207420_at | -0.0858 | 0.0138 | |
| GSE26886 | COLEC10 | 10584 | 207420_at | 0.0859 | 0.3780 | |
| GSE29001 | COLEC10 | 10584 | 207420_at | -0.0766 | 0.6770 | |
| GSE38129 | COLEC10 | 10584 | 207420_at | -0.0087 | 0.9371 | |
| GSE45670 | COLEC10 | 10584 | 207420_at | 0.0953 | 0.3506 | |
| GSE63941 | COLEC10 | 10584 | 207420_at | -1.7944 | 0.0000 | |
| GSE77861 | COLEC10 | 10584 | 207420_at | -0.0960 | 0.4371 | |
| TCGA | COLEC10 | 10584 | RNAseq | 1.2745 | 0.0842 |
Upregulated datasets: 0; Downregulated datasets: 1.
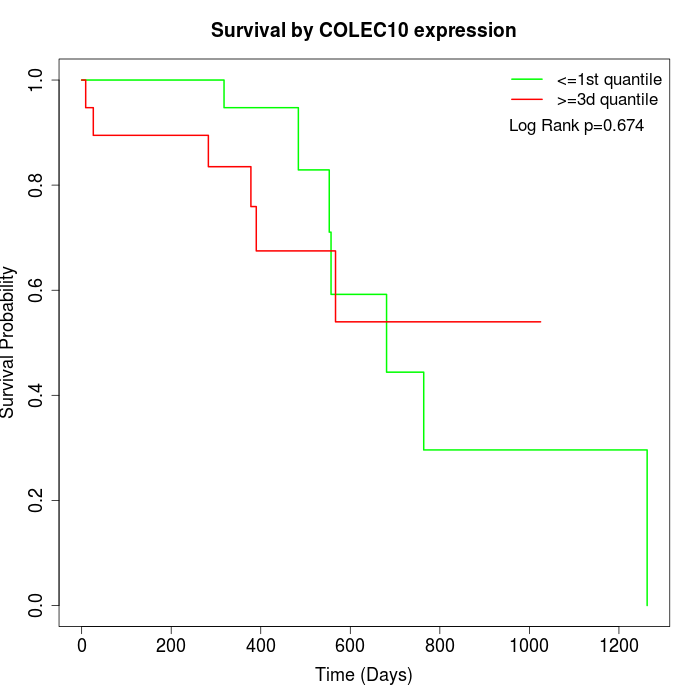
Survival by COLEC10 expression:
Note: Click image to view full size file.
Copy number change of COLEC10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | COLEC10 | 10584 | 19 | 1 | 10 | |
| GSE20123 | COLEC10 | 10584 | 20 | 1 | 9 | |
| GSE43470 | COLEC10 | 10584 | 23 | 0 | 20 | |
| GSE46452 | COLEC10 | 10584 | 26 | 0 | 33 | |
| GSE47630 | COLEC10 | 10584 | 24 | 0 | 16 | |
| GSE54993 | COLEC10 | 10584 | 0 | 20 | 50 | |
| GSE54994 | COLEC10 | 10584 | 37 | 1 | 15 | |
| GSE60625 | COLEC10 | 10584 | 0 | 4 | 7 | |
| GSE74703 | COLEC10 | 10584 | 19 | 0 | 17 | |
| GSE74704 | COLEC10 | 10584 | 13 | 1 | 6 | |
| TCGA | COLEC10 | 10584 | 67 | 2 | 27 |
Total number of gains: 248; Total number of losses: 30; Total Number of normals: 210.
Somatic mutations of COLEC10:
Generating mutation plots.
Highly correlated genes for COLEC10:
Showing top 20/692 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| COLEC10 | FOLH1 | 0.741522 | 4 | 0 | 4 |
| COLEC10 | KCNAB3 | 0.732517 | 5 | 0 | 5 |
| COLEC10 | KIR2DL5A | 0.730752 | 6 | 0 | 5 |
| COLEC10 | SLC17A1 | 0.728763 | 5 | 0 | 5 |
| COLEC10 | INE1 | 0.717276 | 4 | 0 | 3 |
| COLEC10 | PRAMEF12 | 0.716187 | 5 | 0 | 5 |
| COLEC10 | KIF5A | 0.716164 | 6 | 0 | 6 |
| COLEC10 | CPS1-IT1 | 0.7094 | 3 | 0 | 3 |
| COLEC10 | KCNJ10 | 0.706105 | 4 | 0 | 4 |
| COLEC10 | GSTM5 | 0.703983 | 3 | 0 | 3 |
| COLEC10 | BTN1A1 | 0.698927 | 5 | 0 | 5 |
| COLEC10 | GNG3 | 0.698723 | 4 | 0 | 4 |
| COLEC10 | IL1RAPL2 | 0.695881 | 5 | 0 | 5 |
| COLEC10 | TBXA2R | 0.695087 | 3 | 0 | 3 |
| COLEC10 | CYP3A4 | 0.694754 | 4 | 0 | 4 |
| COLEC10 | ZNF771 | 0.69457 | 3 | 0 | 3 |
| COLEC10 | DAB1 | 0.693783 | 5 | 0 | 5 |
| COLEC10 | CCDC87 | 0.692063 | 4 | 0 | 4 |
| COLEC10 | SLC5A4 | 0.688614 | 4 | 0 | 4 |
| COLEC10 | GALNT8 | 0.688031 | 5 | 0 | 5 |
For details and further investigation, click here