| Full name: DAB adaptor protein 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p32.2 | ||
| Entrez ID: 1600 | HGNC ID: HGNC:2661 | Ensembl Gene: ENSG00000173406 | OMIM ID: 603448 |
| Drug and gene relationship at DGIdb | |||
Expression of DAB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DAB1 | 1600 | 220611_at | -0.0236 | 0.9380 | |
| GSE20347 | DAB1 | 1600 | 220611_at | 0.0326 | 0.6928 | |
| GSE23400 | DAB1 | 1600 | 220611_at | -0.1211 | 0.0024 | |
| GSE26886 | DAB1 | 1600 | 220611_at | 0.1159 | 0.2156 | |
| GSE29001 | DAB1 | 1600 | 220611_at | -0.0049 | 0.9803 | |
| GSE38129 | DAB1 | 1600 | 220611_at | -0.0390 | 0.7186 | |
| GSE45670 | DAB1 | 1600 | 228329_at | -0.0780 | 0.8102 | |
| GSE53622 | DAB1 | 1600 | 40820 | 0.5415 | 0.0000 | |
| GSE53624 | DAB1 | 1600 | 40820 | 0.3613 | 0.0000 | |
| GSE63941 | DAB1 | 1600 | 228329_at | -4.3678 | 0.0000 | |
| GSE77861 | DAB1 | 1600 | 220611_at | -0.0775 | 0.3781 | |
| GSE97050 | DAB1 | 1600 | A_23_P23850 | 0.3343 | 0.2541 | |
| SRP133303 | DAB1 | 1600 | RNAseq | 0.7693 | 0.0255 | |
| SRP159526 | DAB1 | 1600 | RNAseq | 0.7385 | 0.1940 | |
| SRP193095 | DAB1 | 1600 | RNAseq | -0.0592 | 0.8496 | |
| TCGA | DAB1 | 1600 | RNAseq | -1.2752 | 0.0401 |
Upregulated datasets: 0; Downregulated datasets: 2.
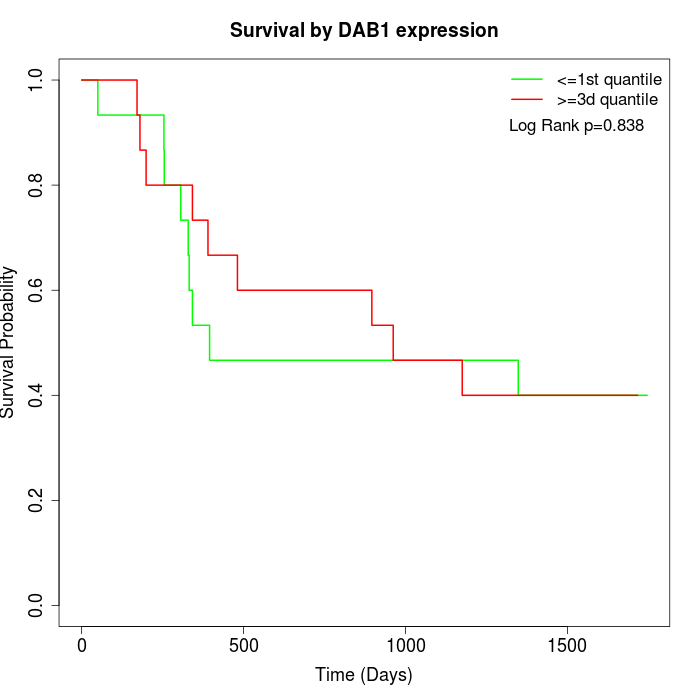
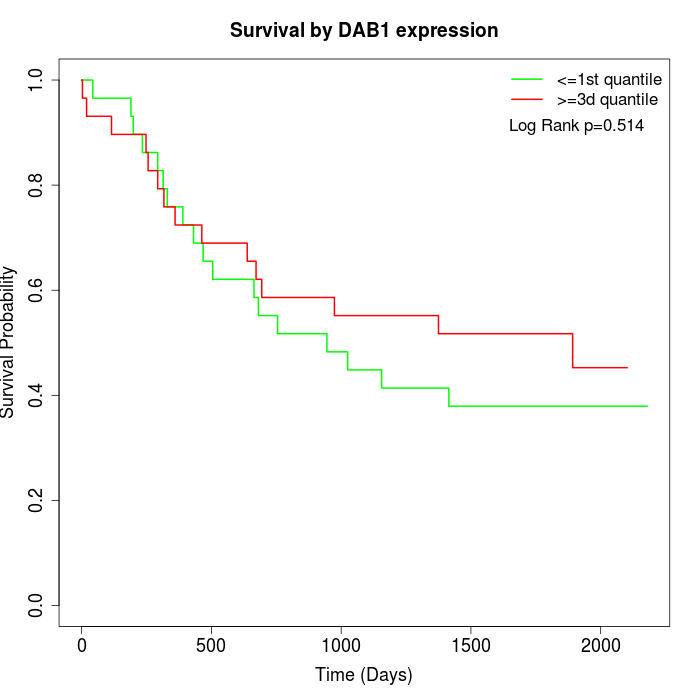
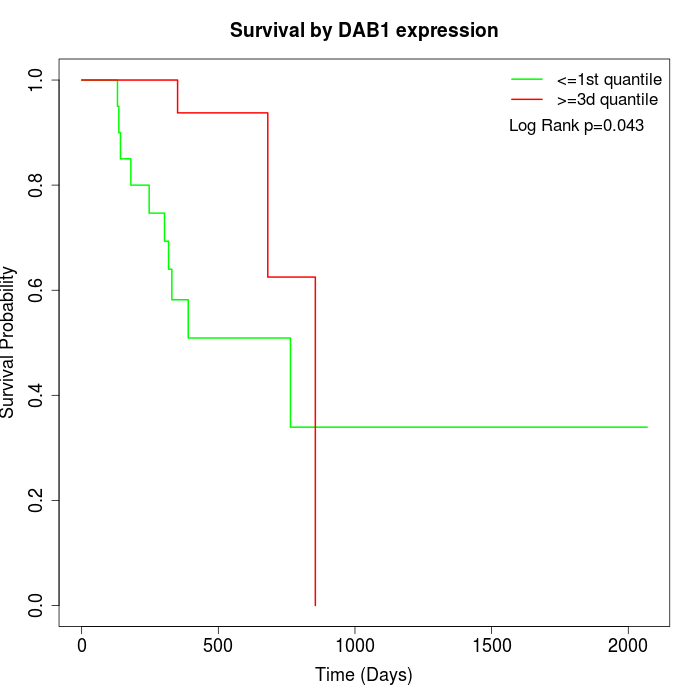
Survival by DAB1 expression:
Note: Click image to view full size file.
Copy number change of DAB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DAB1 | 1600 | 3 | 5 | 22 | |
| GSE20123 | DAB1 | 1600 | 3 | 4 | 23 | |
| GSE43470 | DAB1 | 1600 | 7 | 1 | 35 | |
| GSE46452 | DAB1 | 1600 | 2 | 1 | 56 | |
| GSE47630 | DAB1 | 1600 | 8 | 5 | 27 | |
| GSE54993 | DAB1 | 1600 | 0 | 0 | 70 | |
| GSE54994 | DAB1 | 1600 | 9 | 2 | 42 | |
| GSE60625 | DAB1 | 1600 | 0 | 0 | 11 | |
| GSE74703 | DAB1 | 1600 | 6 | 1 | 29 | |
| GSE74704 | DAB1 | 1600 | 2 | 0 | 18 | |
| TCGA | DAB1 | 1600 | 10 | 19 | 67 |
Total number of gains: 50; Total number of losses: 38; Total Number of normals: 400.
Somatic mutations of DAB1:
Generating mutation plots.
Highly correlated genes for DAB1:
Showing top 20/619 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DAB1 | AQP8 | 0.788807 | 5 | 0 | 5 |
| DAB1 | USH1C | 0.784492 | 5 | 0 | 5 |
| DAB1 | ERN1 | 0.784026 | 4 | 0 | 4 |
| DAB1 | CYP2A7 | 0.781156 | 4 | 0 | 4 |
| DAB1 | G6PC2 | 0.776367 | 4 | 0 | 4 |
| DAB1 | MAG | 0.772285 | 4 | 0 | 4 |
| DAB1 | FOXN3-AS2 | 0.772186 | 4 | 0 | 4 |
| DAB1 | LMAN1L | 0.767202 | 5 | 0 | 5 |
| DAB1 | GABRA5 | 0.762585 | 3 | 0 | 3 |
| DAB1 | PRAMEF12 | 0.761007 | 5 | 0 | 5 |
| DAB1 | CCDC33 | 0.755368 | 5 | 0 | 5 |
| DAB1 | CD28 | 0.754787 | 4 | 0 | 4 |
| DAB1 | OR10H3 | 0.754289 | 4 | 0 | 4 |
| DAB1 | FOLH1 | 0.752426 | 5 | 0 | 5 |
| DAB1 | LINC01140 | 0.732147 | 3 | 0 | 3 |
| DAB1 | ODF1 | 0.730843 | 4 | 0 | 4 |
| DAB1 | USP49 | 0.730109 | 5 | 0 | 5 |
| DAB1 | RNF185-AS1 | 0.728673 | 4 | 0 | 4 |
| DAB1 | RNF17 | 0.728516 | 5 | 0 | 5 |
| DAB1 | CYP3A4 | 0.728333 | 5 | 0 | 5 |
For details and further investigation, click here