| Full name: inactivation escape 1 | Alias Symbol: NCRNA00010 | ||
| Type: non-coding RNA | Cytoband: Xp11.3 | ||
| Entrez ID: 8552 | HGNC ID: HGNC:6060 | Ensembl Gene: ENSG00000224975 | OMIM ID: 300164 |
| Drug and gene relationship at DGIdb | |||
Expression of INE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | INE1 | 8552 | 207252_at | 0.0741 | 0.7862 | |
| GSE20347 | INE1 | 8552 | 207252_at | 0.1088 | 0.2343 | |
| GSE23400 | INE1 | 8552 | 207252_at | -0.0336 | 0.2537 | |
| GSE26886 | INE1 | 8552 | 207252_at | 0.0013 | 0.9905 | |
| GSE29001 | INE1 | 8552 | 207252_at | 0.0206 | 0.9112 | |
| GSE38129 | INE1 | 8552 | 207252_at | -0.0401 | 0.7080 | |
| GSE45670 | INE1 | 8552 | 207252_at | 0.0938 | 0.3415 | |
| GSE53622 | INE1 | 8552 | 59642 | 0.2981 | 0.0061 | |
| GSE53624 | INE1 | 8552 | 59642 | 0.2610 | 0.0087 | |
| GSE63941 | INE1 | 8552 | 207252_at | 0.1099 | 0.4055 | |
| GSE77861 | INE1 | 8552 | 207252_at | -0.0283 | 0.7367 | |
| SRP007169 | INE1 | 8552 | RNAseq | -0.4832 | 0.4949 | |
| SRP133303 | INE1 | 8552 | RNAseq | 0.0494 | 0.7956 | |
| SRP159526 | INE1 | 8552 | RNAseq | 0.4228 | 0.2536 | |
| SRP193095 | INE1 | 8552 | RNAseq | 0.0003 | 0.9987 | |
| SRP219564 | INE1 | 8552 | RNAseq | -0.3654 | 0.3866 | |
| TCGA | INE1 | 8552 | RNAseq | -0.2244 | 0.1887 |
Upregulated datasets: 0; Downregulated datasets: 0.
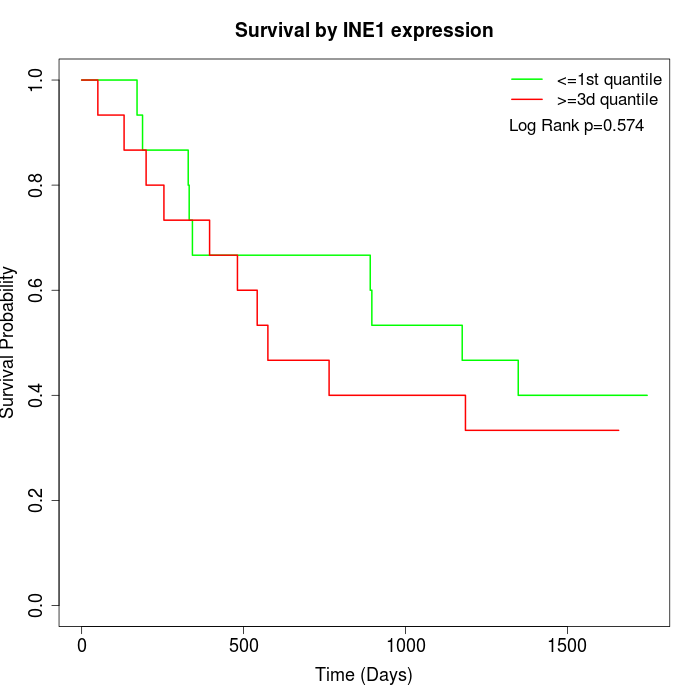
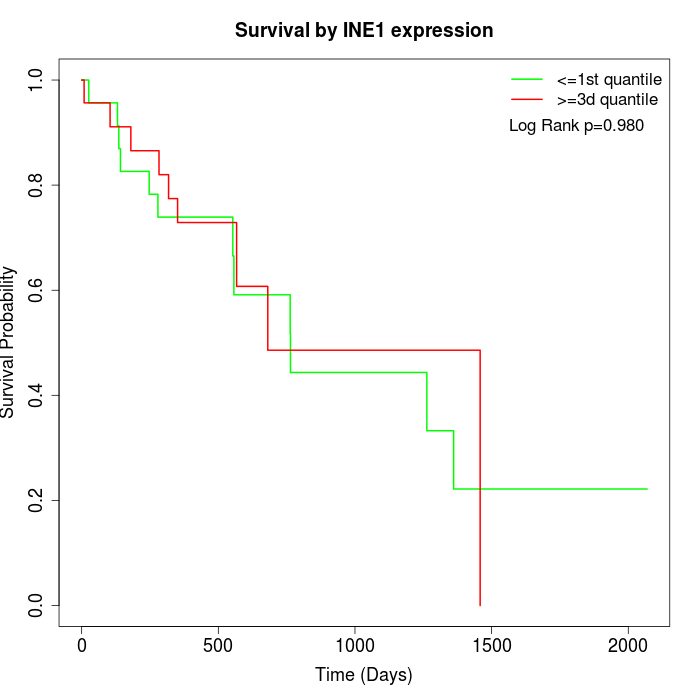
Survival by INE1 expression:
Note: Click image to view full size file.
Copy number change of INE1:
No record found for this gene.
Somatic mutations of INE1:
Generating mutation plots.
Highly correlated genes for INE1:
Showing top 20/672 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| INE1 | SFTPB | 0.766527 | 3 | 0 | 3 |
| INE1 | KIR2DL2 | 0.731585 | 4 | 0 | 4 |
| INE1 | COLEC10 | 0.717276 | 4 | 0 | 3 |
| INE1 | NEUROD2 | 0.705921 | 4 | 0 | 4 |
| INE1 | KCNV1 | 0.705866 | 3 | 0 | 3 |
| INE1 | GDF5 | 0.703578 | 4 | 0 | 4 |
| INE1 | CA7 | 0.700789 | 5 | 0 | 5 |
| INE1 | ADAM30 | 0.693064 | 5 | 0 | 5 |
| INE1 | ZNF236 | 0.691795 | 5 | 0 | 5 |
| INE1 | NRP2 | 0.69106 | 3 | 0 | 3 |
| INE1 | PEAK1 | 0.688605 | 4 | 0 | 4 |
| INE1 | NTRK3 | 0.680061 | 4 | 0 | 4 |
| INE1 | KIR3DL3 | 0.68004 | 5 | 0 | 4 |
| INE1 | PRR34 | 0.680004 | 4 | 0 | 3 |
| INE1 | OR10H1 | 0.676825 | 6 | 0 | 4 |
| INE1 | GML | 0.676527 | 5 | 0 | 5 |
| INE1 | KIR2DS3 | 0.671821 | 5 | 0 | 5 |
| INE1 | KCNIP2 | 0.670855 | 4 | 0 | 4 |
| INE1 | EPN1 | 0.669059 | 3 | 0 | 3 |
| INE1 | MAP3K19 | 0.667885 | 4 | 0 | 3 |
For details and further investigation, click here