| Full name: CMT1A duplicated region transcript 15 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17p12 | ||
| Entrez ID: 146822 | HGNC ID: HGNC:14395 | Ensembl Gene: ENSG00000223510 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CDRT15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CDRT15 | 146822 | 240760_at | -0.0791 | 0.7709 | |
| GSE26886 | CDRT15 | 146822 | 240760_at | 0.3490 | 0.0044 | |
| GSE45670 | CDRT15 | 146822 | 240760_at | 0.1154 | 0.2591 | |
| GSE53622 | CDRT15 | 146822 | 102208 | 0.1083 | 0.1459 | |
| GSE53624 | CDRT15 | 146822 | 7360 | 0.2122 | 0.1682 | |
| GSE63941 | CDRT15 | 146822 | 240760_at | -0.0222 | 0.9159 | |
| GSE77861 | CDRT15 | 146822 | 240760_at | -0.0100 | 0.9568 | |
| GSE97050 | CDRT15 | 146822 | A_24_P756766 | 0.1814 | 0.3638 |
Upregulated datasets: 0; Downregulated datasets: 0.
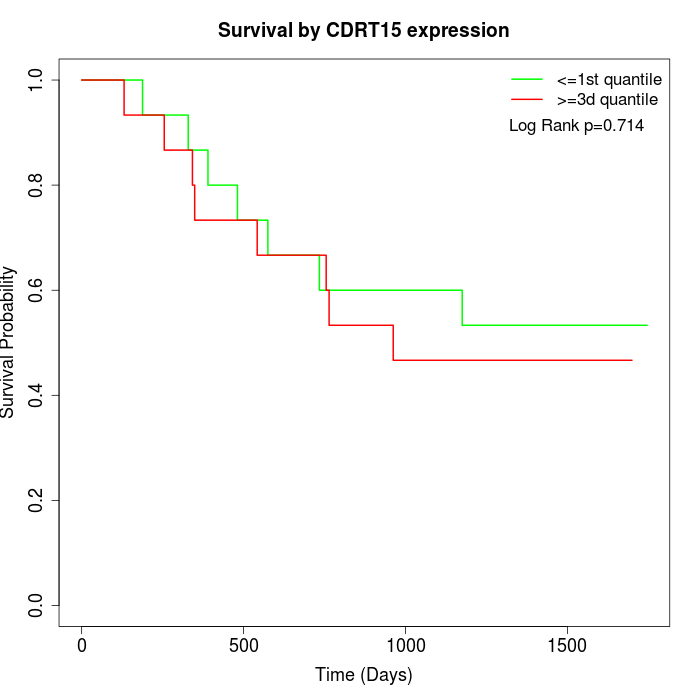
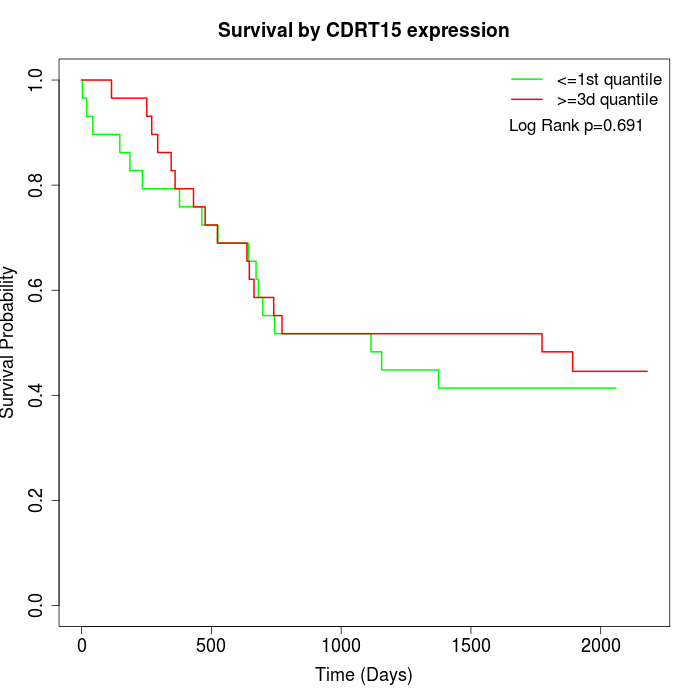
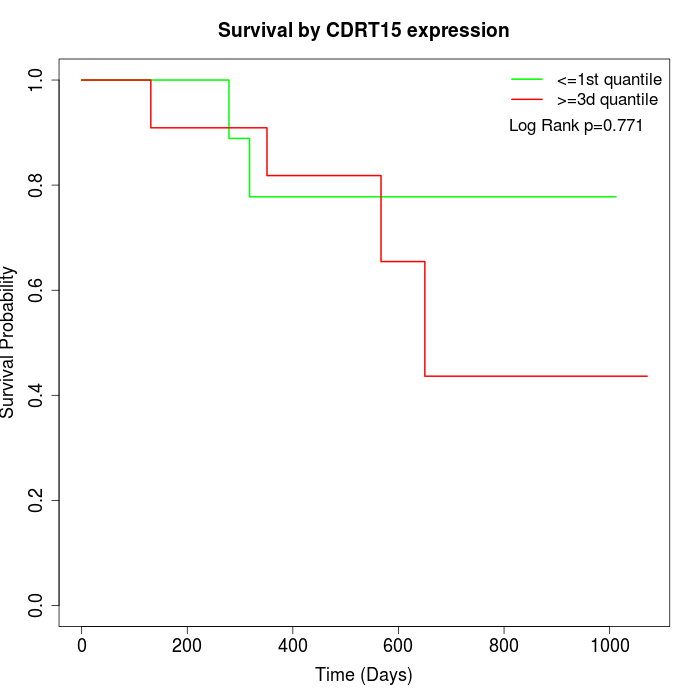
Survival by CDRT15 expression:
Note: Click image to view full size file.
Copy number change of CDRT15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CDRT15 | 146822 | 3 | 3 | 24 | |
| GSE20123 | CDRT15 | 146822 | 3 | 3 | 24 | |
| GSE43470 | CDRT15 | 146822 | 2 | 5 | 36 | |
| GSE46452 | CDRT15 | 146822 | 34 | 1 | 24 | |
| GSE47630 | CDRT15 | 146822 | 7 | 1 | 32 | |
| GSE54993 | CDRT15 | 146822 | 3 | 2 | 65 | |
| GSE54994 | CDRT15 | 146822 | 6 | 6 | 41 | |
| GSE60625 | CDRT15 | 146822 | 4 | 0 | 7 | |
| GSE74703 | CDRT15 | 146822 | 2 | 4 | 30 | |
| GSE74704 | CDRT15 | 146822 | 2 | 2 | 16 | |
| TCGA | CDRT15 | 146822 | 18 | 22 | 56 |
Total number of gains: 84; Total number of losses: 49; Total Number of normals: 355.
Somatic mutations of CDRT15:
Generating mutation plots.
Highly correlated genes for CDRT15:
Showing top 20/486 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CDRT15 | STRC | 0.792319 | 3 | 0 | 3 |
| CDRT15 | C19orf81 | 0.775033 | 3 | 0 | 3 |
| CDRT15 | OR4D6 | 0.771122 | 3 | 0 | 3 |
| CDRT15 | DEFB128 | 0.751839 | 3 | 0 | 3 |
| CDRT15 | SIM1 | 0.746392 | 4 | 0 | 4 |
| CDRT15 | CLCN2 | 0.74633 | 3 | 0 | 3 |
| CDRT15 | HYAL1 | 0.744514 | 3 | 0 | 3 |
| CDRT15 | MYL7 | 0.744073 | 5 | 0 | 4 |
| CDRT15 | PIP5KL1 | 0.743052 | 3 | 0 | 3 |
| CDRT15 | OR4D1 | 0.742236 | 4 | 0 | 3 |
| CDRT15 | PDIA2 | 0.741277 | 3 | 0 | 3 |
| CDRT15 | RHBDL1 | 0.732047 | 3 | 0 | 3 |
| CDRT15 | CHST13 | 0.731364 | 3 | 0 | 3 |
| CDRT15 | RHEBL1 | 0.730815 | 3 | 0 | 3 |
| CDRT15 | PLA2G2C | 0.730203 | 3 | 0 | 3 |
| CDRT15 | IL20 | 0.729216 | 3 | 0 | 3 |
| CDRT15 | ZFPM1 | 0.728315 | 3 | 0 | 3 |
| CDRT15 | ZGLP1 | 0.725619 | 3 | 0 | 3 |
| CDRT15 | PTPRN | 0.723224 | 3 | 0 | 3 |
| CDRT15 | C9orf153 | 0.722706 | 3 | 0 | 3 |
For details and further investigation, click here