| Full name: DC-STAMP domain containing 1 | Alias Symbol: FLJ32785 | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 149095 | HGNC ID: HGNC:26539 | Ensembl Gene: ENSG00000163357 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of DCST1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DCST1 | 149095 | 1553860_at | -0.1448 | 0.7045 | |
| GSE26886 | DCST1 | 149095 | 1553860_at | -0.1595 | 0.3776 | |
| GSE45670 | DCST1 | 149095 | 1553860_at | 0.0427 | 0.7115 | |
| GSE53622 | DCST1 | 149095 | 19636 | 0.0528 | 0.6607 | |
| GSE53624 | DCST1 | 149095 | 19636 | -0.2540 | 0.0380 | |
| GSE63941 | DCST1 | 149095 | 1553860_at | 0.5410 | 0.0159 | |
| GSE77861 | DCST1 | 149095 | 1553860_at | -0.3261 | 0.0262 | |
| SRP133303 | DCST1 | 149095 | RNAseq | -0.2447 | 0.3535 | |
| SRP159526 | DCST1 | 149095 | RNAseq | -0.2872 | 0.6500 | |
| TCGA | DCST1 | 149095 | RNAseq | 1.5014 | 0.0789 |
Upregulated datasets: 0; Downregulated datasets: 0.
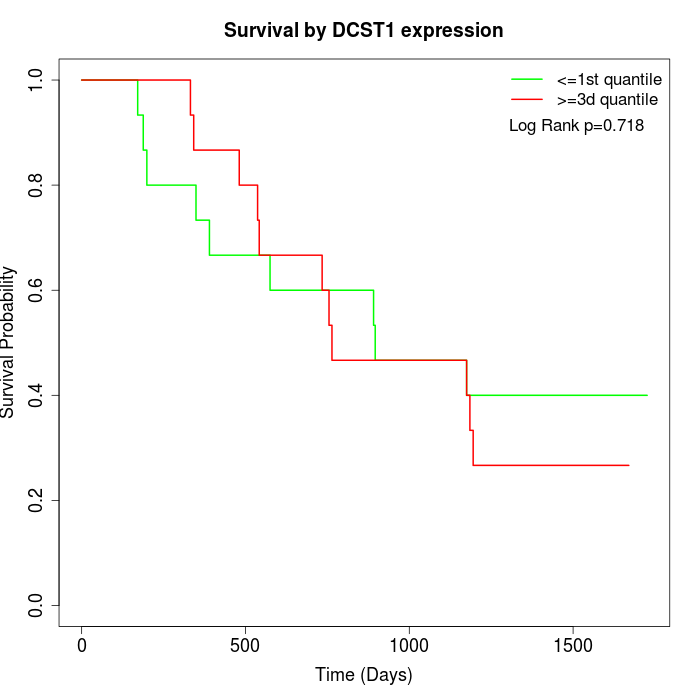
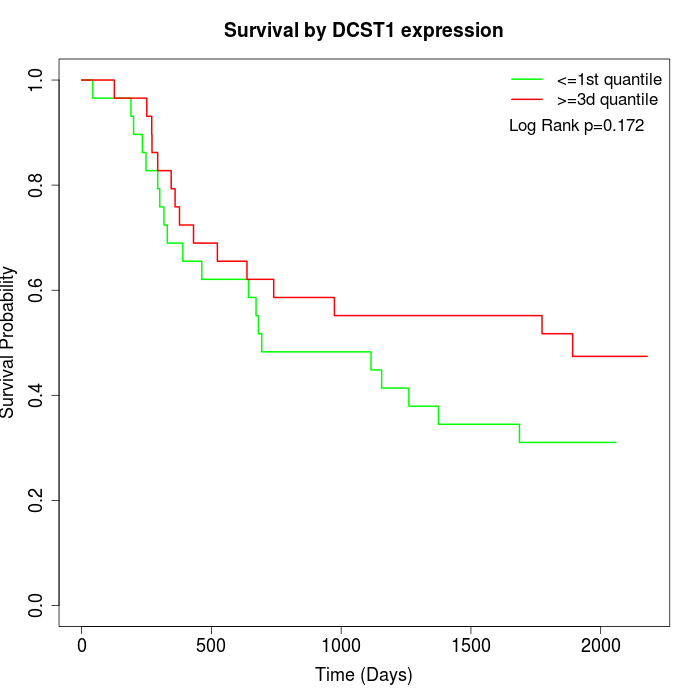
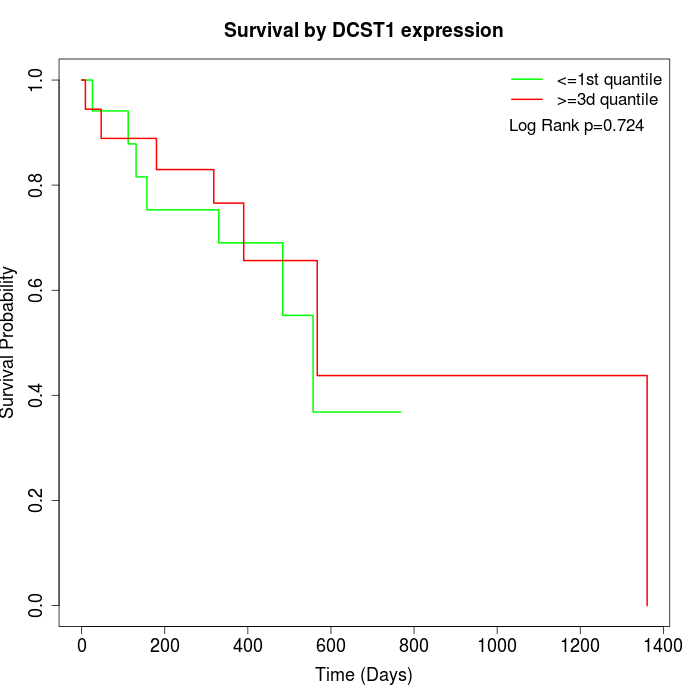
Survival by DCST1 expression:
Note: Click image to view full size file.
Copy number change of DCST1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DCST1 | 149095 | 14 | 0 | 16 | |
| GSE20123 | DCST1 | 149095 | 13 | 0 | 17 | |
| GSE43470 | DCST1 | 149095 | 6 | 2 | 35 | |
| GSE46452 | DCST1 | 149095 | 2 | 1 | 56 | |
| GSE47630 | DCST1 | 149095 | 14 | 0 | 26 | |
| GSE54993 | DCST1 | 149095 | 0 | 4 | 66 | |
| GSE54994 | DCST1 | 149095 | 16 | 0 | 37 | |
| GSE60625 | DCST1 | 149095 | 0 | 0 | 11 | |
| GSE74703 | DCST1 | 149095 | 6 | 1 | 29 | |
| GSE74704 | DCST1 | 149095 | 6 | 0 | 14 | |
| TCGA | DCST1 | 149095 | 38 | 2 | 56 |
Total number of gains: 115; Total number of losses: 10; Total Number of normals: 363.
Somatic mutations of DCST1:
Generating mutation plots.
Highly correlated genes for DCST1:
Showing top 20/238 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DCST1 | VGLL2 | 0.772629 | 3 | 0 | 3 |
| DCST1 | C10orf90 | 0.707662 | 3 | 0 | 3 |
| DCST1 | LRRC3 | 0.69855 | 3 | 0 | 3 |
| DCST1 | AMACR | 0.676263 | 3 | 0 | 3 |
| DCST1 | C10orf62 | 0.67329 | 4 | 0 | 3 |
| DCST1 | ACSBG1 | 0.672667 | 3 | 0 | 3 |
| DCST1 | ASB12 | 0.671282 | 3 | 0 | 3 |
| DCST1 | MMP17 | 0.666437 | 3 | 0 | 3 |
| DCST1 | BTNL3 | 0.665654 | 3 | 0 | 3 |
| DCST1 | NEUROD6 | 0.655599 | 3 | 0 | 3 |
| DCST1 | MYOZ1 | 0.655214 | 3 | 0 | 3 |
| DCST1 | CNR2 | 0.654428 | 4 | 0 | 3 |
| DCST1 | ENPP3 | 0.645975 | 4 | 0 | 4 |
| DCST1 | MAPT-AS1 | 0.642622 | 5 | 0 | 4 |
| DCST1 | OPN5 | 0.64141 | 3 | 0 | 3 |
| DCST1 | MIOX | 0.63864 | 3 | 0 | 3 |
| DCST1 | NXNL1 | 0.634842 | 3 | 0 | 3 |
| DCST1 | FCER2 | 0.632652 | 3 | 0 | 3 |
| DCST1 | ETV2 | 0.630387 | 3 | 0 | 3 |
| DCST1 | SLC6A18 | 0.62984 | 3 | 0 | 3 |
For details and further investigation, click here