| Full name: 2,4-dienoyl-CoA reductase 2 | Alias Symbol: PDCR|SDR17C1 | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 26063 | HGNC ID: HGNC:2754 | Ensembl Gene: ENSG00000242612 | OMIM ID: 615839 |
| Drug and gene relationship at DGIdb | |||
Expression of DECR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DECR2 | 26063 | 219664_s_at | -0.2204 | 0.5183 | |
| GSE20347 | DECR2 | 26063 | 219664_s_at | -0.0909 | 0.4389 | |
| GSE23400 | DECR2 | 26063 | 219664_s_at | -0.0959 | 0.0319 | |
| GSE26886 | DECR2 | 26063 | 219664_s_at | 0.6749 | 0.0000 | |
| GSE29001 | DECR2 | 26063 | 219664_s_at | -0.1000 | 0.6204 | |
| GSE38129 | DECR2 | 26063 | 219664_s_at | -0.1424 | 0.1513 | |
| GSE45670 | DECR2 | 26063 | 219664_s_at | -0.0124 | 0.9229 | |
| GSE53622 | DECR2 | 26063 | 15592 | -0.1983 | 0.0017 | |
| GSE53624 | DECR2 | 26063 | 15592 | -0.0889 | 0.2058 | |
| GSE63941 | DECR2 | 26063 | 219664_s_at | -0.5090 | 0.1426 | |
| GSE77861 | DECR2 | 26063 | 219664_s_at | -0.1193 | 0.5731 | |
| GSE97050 | DECR2 | 26063 | A_33_P3393408 | -0.0810 | 0.7163 | |
| SRP007169 | DECR2 | 26063 | RNAseq | -1.6069 | 0.0017 | |
| SRP008496 | DECR2 | 26063 | RNAseq | -1.2579 | 0.0006 | |
| SRP064894 | DECR2 | 26063 | RNAseq | 0.2412 | 0.3819 | |
| SRP133303 | DECR2 | 26063 | RNAseq | -0.0275 | 0.9204 | |
| SRP159526 | DECR2 | 26063 | RNAseq | -0.1671 | 0.6529 | |
| SRP193095 | DECR2 | 26063 | RNAseq | -0.5633 | 0.0023 | |
| SRP219564 | DECR2 | 26063 | RNAseq | -0.1041 | 0.8151 | |
| TCGA | DECR2 | 26063 | RNAseq | -0.0763 | 0.3370 |
Upregulated datasets: 0; Downregulated datasets: 2.
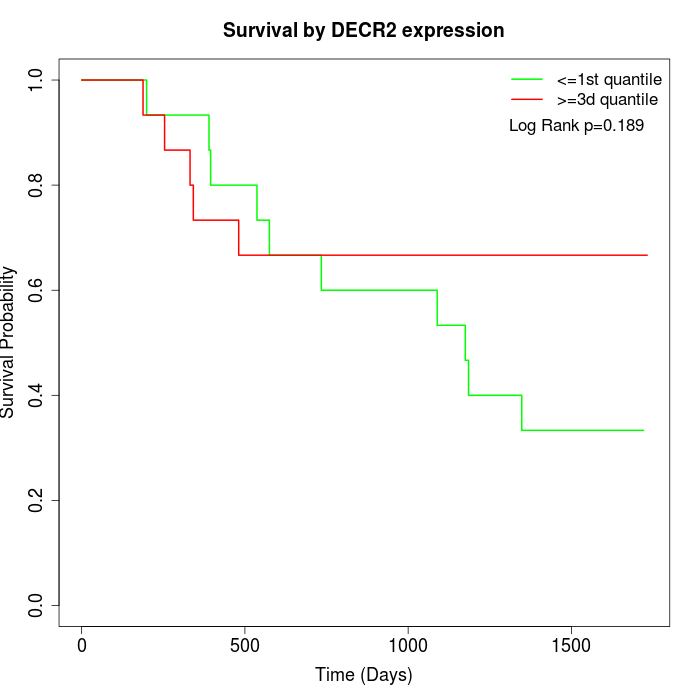
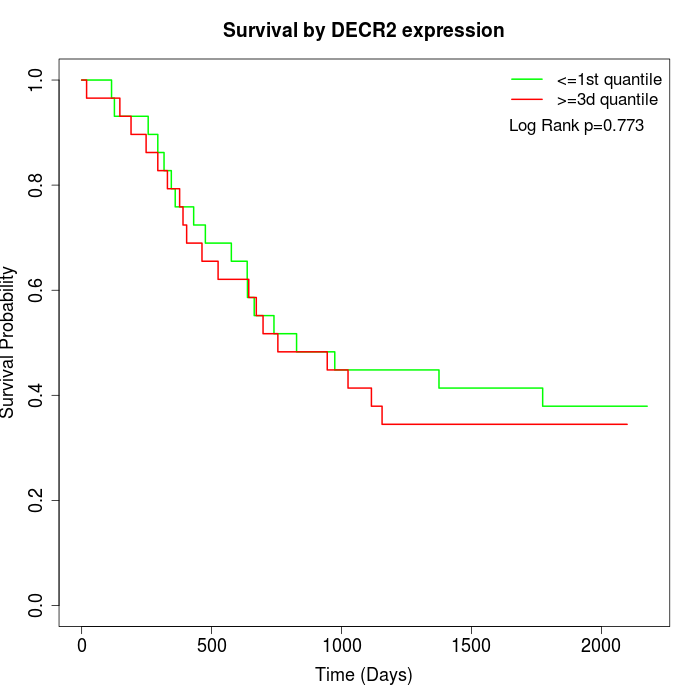
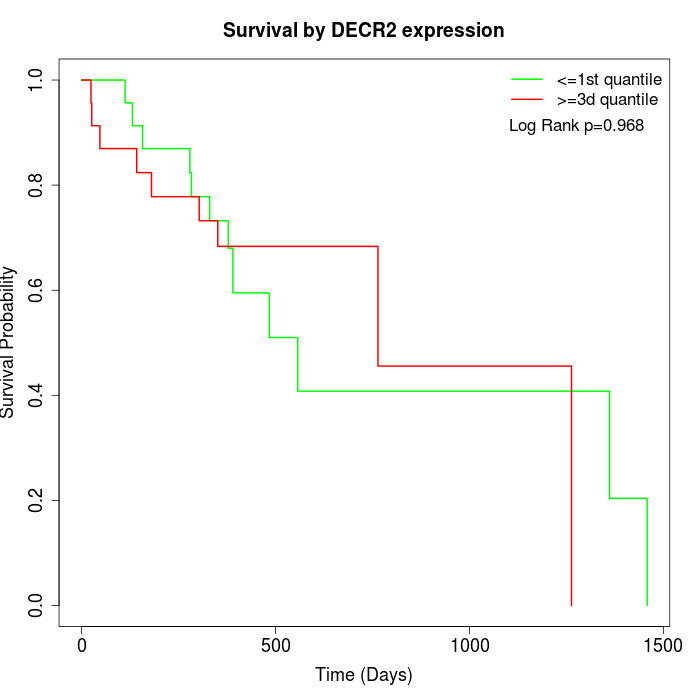
Survival by DECR2 expression:
Note: Click image to view full size file.
Copy number change of DECR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DECR2 | 26063 | 5 | 5 | 20 | |
| GSE20123 | DECR2 | 26063 | 5 | 4 | 21 | |
| GSE43470 | DECR2 | 26063 | 3 | 7 | 33 | |
| GSE46452 | DECR2 | 26063 | 38 | 1 | 20 | |
| GSE47630 | DECR2 | 26063 | 13 | 6 | 21 | |
| GSE54993 | DECR2 | 26063 | 3 | 5 | 62 | |
| GSE54994 | DECR2 | 26063 | 4 | 10 | 39 | |
| GSE60625 | DECR2 | 26063 | 4 | 0 | 7 | |
| GSE74703 | DECR2 | 26063 | 3 | 5 | 28 | |
| GSE74704 | DECR2 | 26063 | 3 | 2 | 15 | |
| TCGA | DECR2 | 26063 | 20 | 15 | 61 |
Total number of gains: 101; Total number of losses: 60; Total Number of normals: 327.
Somatic mutations of DECR2:
Generating mutation plots.
Highly correlated genes for DECR2:
Showing top 20/172 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DECR2 | SLC43A2 | 0.765745 | 3 | 0 | 3 |
| DECR2 | DZIP1L | 0.762463 | 3 | 0 | 3 |
| DECR2 | ACYP1 | 0.740526 | 3 | 0 | 3 |
| DECR2 | FAM204A | 0.73233 | 3 | 0 | 3 |
| DECR2 | ANKRD9 | 0.730299 | 3 | 0 | 3 |
| DECR2 | SDR39U1 | 0.723288 | 3 | 0 | 3 |
| DECR2 | ZBTB8OS | 0.71053 | 3 | 0 | 3 |
| DECR2 | MYOF | 0.708832 | 3 | 0 | 3 |
| DECR2 | LZTS1 | 0.706503 | 3 | 0 | 3 |
| DECR2 | BECN1 | 0.692751 | 3 | 0 | 3 |
| DECR2 | E4F1 | 0.68992 | 4 | 0 | 3 |
| DECR2 | DAPK3 | 0.686431 | 3 | 0 | 3 |
| DECR2 | TSPYL2 | 0.68429 | 3 | 0 | 3 |
| DECR2 | DYNC2H1 | 0.683164 | 3 | 0 | 3 |
| DECR2 | PGLS | 0.676911 | 4 | 0 | 3 |
| DECR2 | NCLN | 0.670089 | 3 | 0 | 3 |
| DECR2 | MEGF6 | 0.666484 | 3 | 0 | 3 |
| DECR2 | SLC2A4RG | 0.666444 | 3 | 0 | 3 |
| DECR2 | PRKACA | 0.665288 | 3 | 0 | 3 |
| DECR2 | AKAP8 | 0.664174 | 4 | 0 | 3 |
For details and further investigation, click here