| Full name: dynein axonemal heavy chain 10 | Alias Symbol: FLJ43808 | ||
| Type: protein-coding gene | Cytoband: 12q24.31 | ||
| Entrez ID: 196385 | HGNC ID: HGNC:2941 | Ensembl Gene: ENSG00000197653 | OMIM ID: 605884 |
| Drug and gene relationship at DGIdb | |||
Expression of DNAH10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DNAH10 | 196385 | 1562462_at | -0.0570 | 0.8800 | |
| GSE26886 | DNAH10 | 196385 | 1562462_at | 0.0676 | 0.5466 | |
| GSE45670 | DNAH10 | 196385 | 1562462_at | 0.0664 | 0.5889 | |
| GSE53622 | DNAH10 | 196385 | 79217 | 0.5259 | 0.0003 | |
| GSE53624 | DNAH10 | 196385 | 52370 | 0.2045 | 0.0115 | |
| GSE63941 | DNAH10 | 196385 | 1562462_at | 0.3860 | 0.0448 | |
| GSE77861 | DNAH10 | 196385 | 1562462_at | -0.2839 | 0.0305 | |
| GSE97050 | DNAH10 | 196385 | A_32_P228268 | -0.1552 | 0.6149 | |
| SRP133303 | DNAH10 | 196385 | RNAseq | 0.2446 | 0.3807 | |
| SRP159526 | DNAH10 | 196385 | RNAseq | 1.1278 | 0.0202 | |
| SRP219564 | DNAH10 | 196385 | RNAseq | 0.9485 | 0.1194 | |
| TCGA | DNAH10 | 196385 | RNAseq | -0.7589 | 0.0029 |
Upregulated datasets: 1; Downregulated datasets: 0.
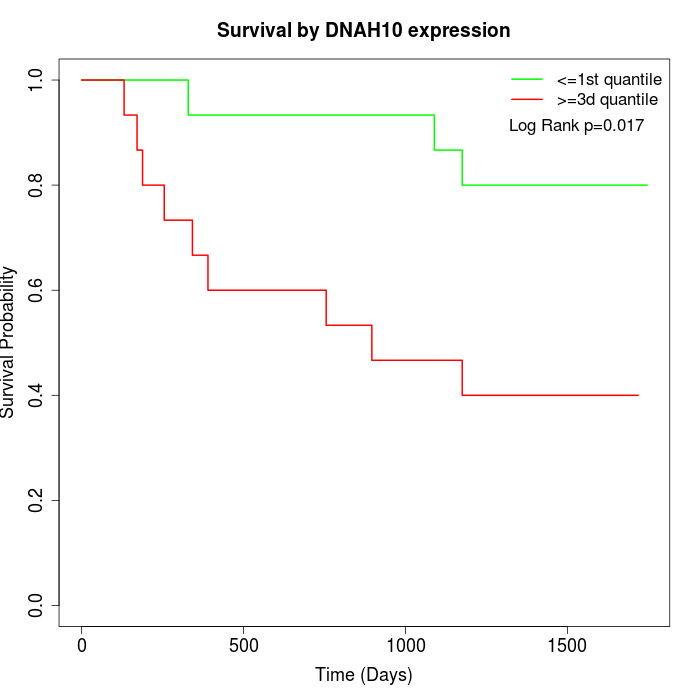
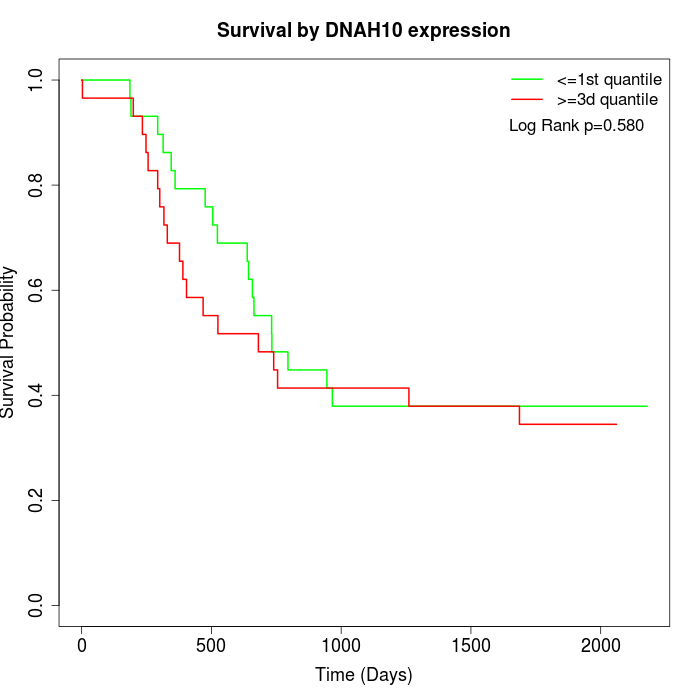
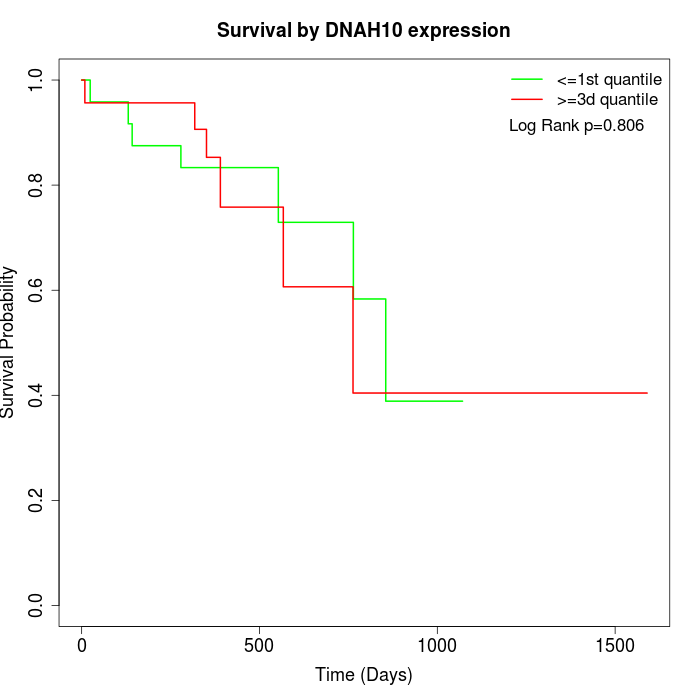
Survival by DNAH10 expression:
Note: Click image to view full size file.
Copy number change of DNAH10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DNAH10 | 196385 | 5 | 5 | 20 | |
| GSE20123 | DNAH10 | 196385 | 5 | 5 | 20 | |
| GSE43470 | DNAH10 | 196385 | 2 | 0 | 41 | |
| GSE46452 | DNAH10 | 196385 | 9 | 1 | 49 | |
| GSE47630 | DNAH10 | 196385 | 9 | 3 | 28 | |
| GSE54993 | DNAH10 | 196385 | 0 | 5 | 65 | |
| GSE54994 | DNAH10 | 196385 | 4 | 8 | 41 | |
| GSE60625 | DNAH10 | 196385 | 0 | 0 | 11 | |
| GSE74703 | DNAH10 | 196385 | 2 | 0 | 34 | |
| GSE74704 | DNAH10 | 196385 | 3 | 3 | 14 | |
| TCGA | DNAH10 | 196385 | 21 | 11 | 64 |
Total number of gains: 60; Total number of losses: 41; Total Number of normals: 387.
Somatic mutations of DNAH10:
Generating mutation plots.
Highly correlated genes for DNAH10:
Showing top 20/69 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DNAH10 | FCN3 | 0.751622 | 3 | 0 | 3 |
| DNAH10 | GZMM | 0.735905 | 3 | 0 | 3 |
| DNAH10 | SLC26A1 | 0.733875 | 3 | 0 | 3 |
| DNAH10 | TRIM46 | 0.713029 | 3 | 0 | 3 |
| DNAH10 | CIDEA | 0.706453 | 3 | 0 | 3 |
| DNAH10 | SEMA3B | 0.698246 | 3 | 0 | 3 |
| DNAH10 | GALR2 | 0.691687 | 3 | 0 | 3 |
| DNAH10 | CABP1 | 0.691285 | 3 | 0 | 3 |
| DNAH10 | C1QTNF6 | 0.684293 | 3 | 0 | 3 |
| DNAH10 | EBP | 0.680993 | 3 | 0 | 3 |
| DNAH10 | ZNF843 | 0.677424 | 3 | 0 | 3 |
| DNAH10 | PGLYRP2 | 0.666185 | 3 | 0 | 3 |
| DNAH10 | CABP7 | 0.664119 | 3 | 0 | 3 |
| DNAH10 | LRRC30 | 0.658047 | 3 | 0 | 3 |
| DNAH10 | FGF21 | 0.653763 | 3 | 0 | 3 |
| DNAH10 | CLEC1B | 0.6515 | 3 | 0 | 3 |
| DNAH10 | TMEM37 | 0.643988 | 5 | 0 | 4 |
| DNAH10 | TMEM213 | 0.641717 | 3 | 0 | 3 |
| DNAH10 | PAX8 | 0.641552 | 4 | 0 | 3 |
| DNAH10 | CRTAC1 | 0.636819 | 4 | 0 | 3 |
For details and further investigation, click here