| Full name: C-type lectin domain family 1 member B | Alias Symbol: CLEC2 | ||
| Type: protein-coding gene | Cytoband: 12p13.31-p13.2 | ||
| Entrez ID: 51266 | HGNC ID: HGNC:24356 | Ensembl Gene: ENSG00000165682 | OMIM ID: 606783 |
| Drug and gene relationship at DGIdb | |||
Expression of CLEC1B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLEC1B | 51266 | 220496_at | -0.0248 | 0.9363 | |
| GSE20347 | CLEC1B | 51266 | 220496_at | -0.0001 | 0.9989 | |
| GSE23400 | CLEC1B | 51266 | 220496_at | -0.1144 | 0.0010 | |
| GSE26886 | CLEC1B | 51266 | 220496_at | -0.0961 | 0.4002 | |
| GSE29001 | CLEC1B | 51266 | 220496_at | -0.0478 | 0.8204 | |
| GSE38129 | CLEC1B | 51266 | 220496_at | -0.1209 | 0.0393 | |
| GSE45670 | CLEC1B | 51266 | 220496_at | 0.0687 | 0.3991 | |
| GSE53622 | CLEC1B | 51266 | 36825 | 0.0570 | 0.5722 | |
| GSE53624 | CLEC1B | 51266 | 36825 | 0.1671 | 0.0437 | |
| GSE63941 | CLEC1B | 51266 | 220496_at | 0.0440 | 0.7553 | |
| GSE77861 | CLEC1B | 51266 | 220496_at | -0.1154 | 0.1589 | |
| GSE97050 | CLEC1B | 51266 | A_33_P3236065 | -0.0568 | 0.8159 |
Upregulated datasets: 0; Downregulated datasets: 0.
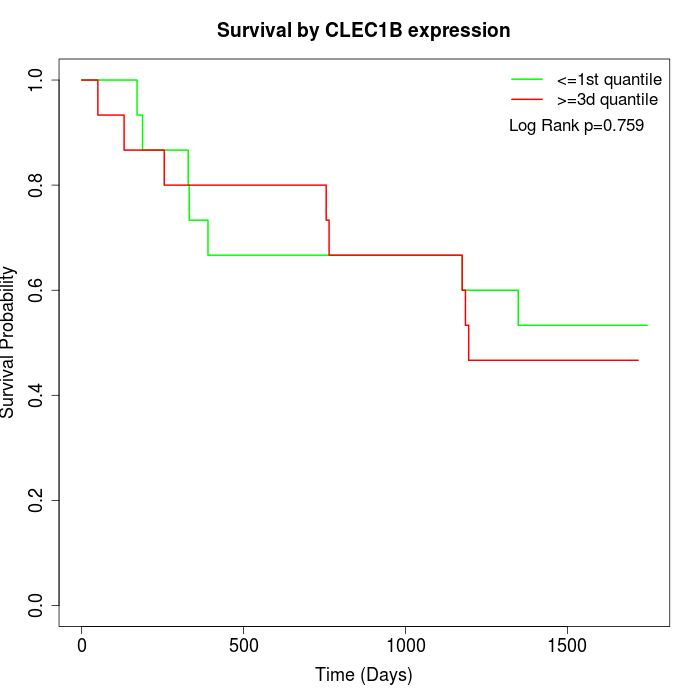
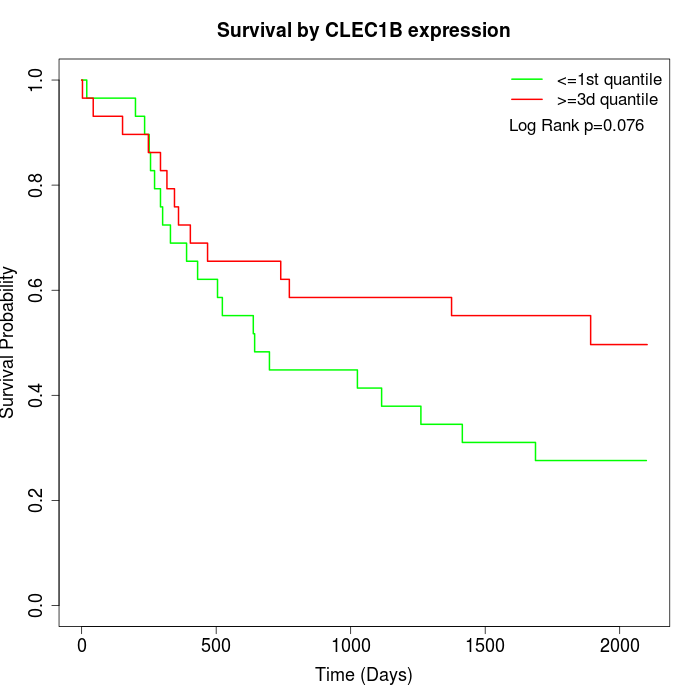
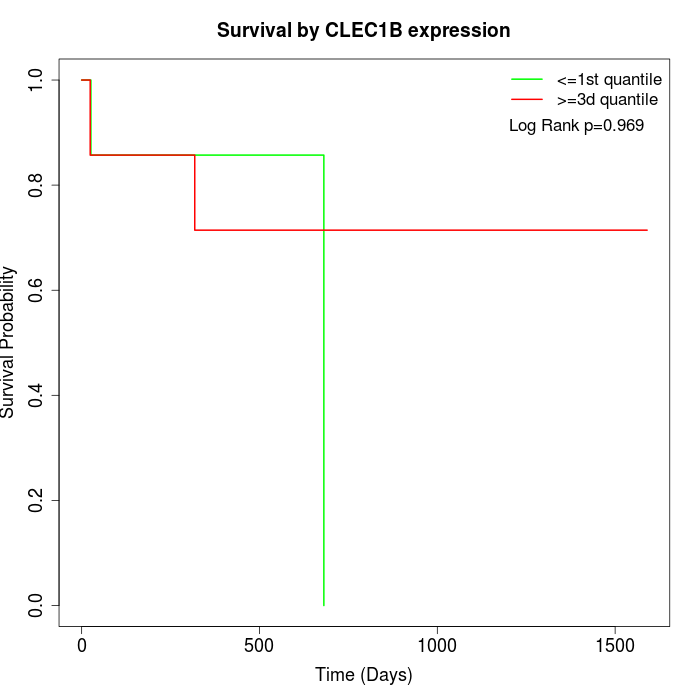
Survival by CLEC1B expression:
Note: Click image to view full size file.
Copy number change of CLEC1B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLEC1B | 51266 | 5 | 3 | 22 | |
| GSE20123 | CLEC1B | 51266 | 5 | 3 | 22 | |
| GSE43470 | CLEC1B | 51266 | 8 | 3 | 32 | |
| GSE46452 | CLEC1B | 51266 | 10 | 1 | 48 | |
| GSE47630 | CLEC1B | 51266 | 14 | 2 | 24 | |
| GSE54993 | CLEC1B | 51266 | 1 | 10 | 59 | |
| GSE54994 | CLEC1B | 51266 | 9 | 2 | 42 | |
| GSE60625 | CLEC1B | 51266 | 0 | 1 | 10 | |
| GSE74703 | CLEC1B | 51266 | 8 | 2 | 26 | |
| GSE74704 | CLEC1B | 51266 | 3 | 2 | 15 | |
| TCGA | CLEC1B | 51266 | 40 | 6 | 50 |
Total number of gains: 103; Total number of losses: 35; Total Number of normals: 350.
Somatic mutations of CLEC1B:
Generating mutation plots.
Highly correlated genes for CLEC1B:
Showing top 20/1190 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLEC1B | PRSS38 | 0.849796 | 3 | 0 | 3 |
| CLEC1B | OR2D2 | 0.793941 | 3 | 0 | 3 |
| CLEC1B | OTOG | 0.792571 | 3 | 0 | 3 |
| CLEC1B | OR4A15 | 0.789055 | 3 | 0 | 3 |
| CLEC1B | FFAR1 | 0.788577 | 4 | 0 | 4 |
| CLEC1B | OR2B11 | 0.787048 | 3 | 0 | 3 |
| CLEC1B | RBM43 | 0.7835 | 3 | 0 | 3 |
| CLEC1B | YY2 | 0.776987 | 3 | 0 | 3 |
| CLEC1B | PROCA1 | 0.768145 | 5 | 0 | 5 |
| CLEC1B | SLC39A5 | 0.759615 | 3 | 0 | 3 |
| CLEC1B | PLA2G2C | 0.7562 | 3 | 0 | 3 |
| CLEC1B | PAX6 | 0.753148 | 3 | 0 | 3 |
| CLEC1B | OR10AD1 | 0.750988 | 3 | 0 | 3 |
| CLEC1B | PRAMEF5 | 0.750671 | 3 | 0 | 3 |
| CLEC1B | STAC2 | 0.738307 | 3 | 0 | 3 |
| CLEC1B | KRTAP10-12 | 0.737512 | 3 | 0 | 3 |
| CLEC1B | MRPS16 | 0.73502 | 3 | 0 | 3 |
| CLEC1B | LYZL2 | 0.733597 | 3 | 0 | 3 |
| CLEC1B | OR2T1 | 0.733173 | 3 | 0 | 3 |
| CLEC1B | SHANK1 | 0.732518 | 5 | 0 | 5 |
For details and further investigation, click here