| Full name: endo-beta-N-acetylglucosaminidase | Alias Symbol: FLJ21865 | ||
| Type: protein-coding gene | Cytoband: 17q25.3 | ||
| Entrez ID: 64772 | HGNC ID: HGNC:24622 | Ensembl Gene: ENSG00000167280 | OMIM ID: 611898 |
| Drug and gene relationship at DGIdb | |||
Expression of ENGASE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ENGASE | 64772 | 65635_at | -0.0711 | 0.8288 | |
| GSE20347 | ENGASE | 64772 | 65635_at | 0.0386 | 0.7646 | |
| GSE23400 | ENGASE | 64772 | 65635_at | 0.0877 | 0.0051 | |
| GSE26886 | ENGASE | 64772 | 65635_at | -0.0267 | 0.8343 | |
| GSE29001 | ENGASE | 64772 | 220349_s_at | 0.6719 | 0.0665 | |
| GSE38129 | ENGASE | 64772 | 65635_at | -0.0841 | 0.4646 | |
| GSE45670 | ENGASE | 64772 | 220349_s_at | 0.2864 | 0.3883 | |
| GSE53622 | ENGASE | 64772 | 18651 | 0.3971 | 0.0000 | |
| GSE53624 | ENGASE | 64772 | 26308 | 0.1293 | 0.1090 | |
| GSE63941 | ENGASE | 64772 | 65635_at | 0.3046 | 0.0713 | |
| GSE77861 | ENGASE | 64772 | 65635_at | 0.0525 | 0.5915 | |
| GSE97050 | ENGASE | 64772 | A_33_P3348220 | 0.0848 | 0.7057 | |
| SRP007169 | ENGASE | 64772 | RNAseq | 0.6859 | 0.1614 | |
| SRP008496 | ENGASE | 64772 | RNAseq | 1.5218 | 0.0002 | |
| SRP064894 | ENGASE | 64772 | RNAseq | 0.5517 | 0.0008 | |
| SRP133303 | ENGASE | 64772 | RNAseq | 0.1217 | 0.3809 | |
| SRP159526 | ENGASE | 64772 | RNAseq | 0.5164 | 0.1778 | |
| SRP193095 | ENGASE | 64772 | RNAseq | 0.7649 | 0.0002 | |
| SRP219564 | ENGASE | 64772 | RNAseq | 0.1656 | 0.5245 | |
| TCGA | ENGASE | 64772 | RNAseq | -0.0561 | 0.4566 |
Upregulated datasets: 1; Downregulated datasets: 0.
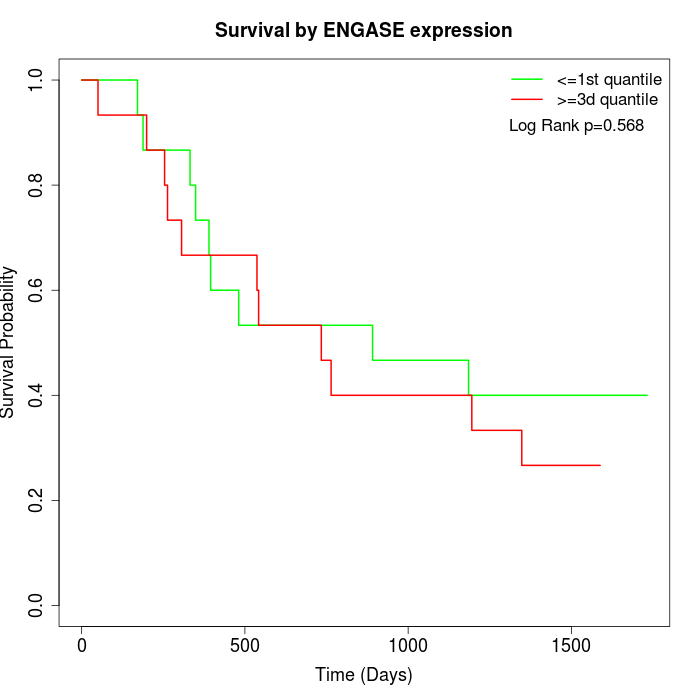
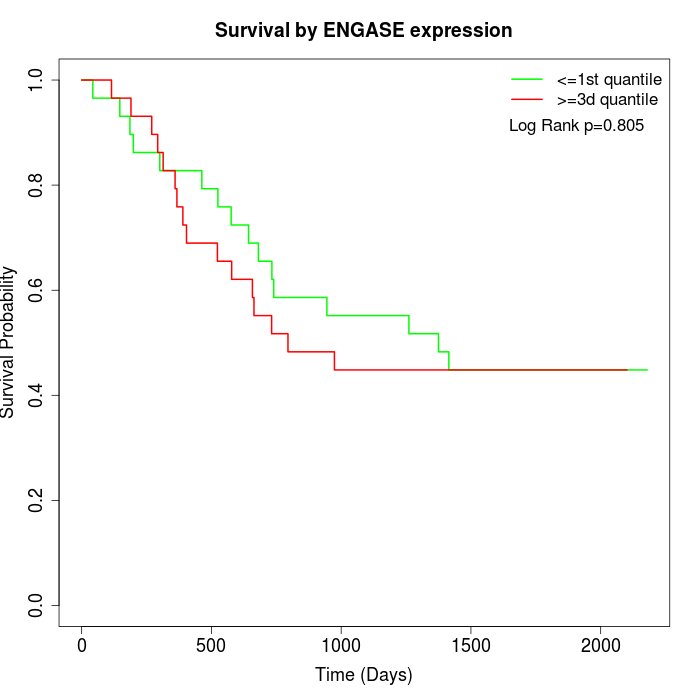
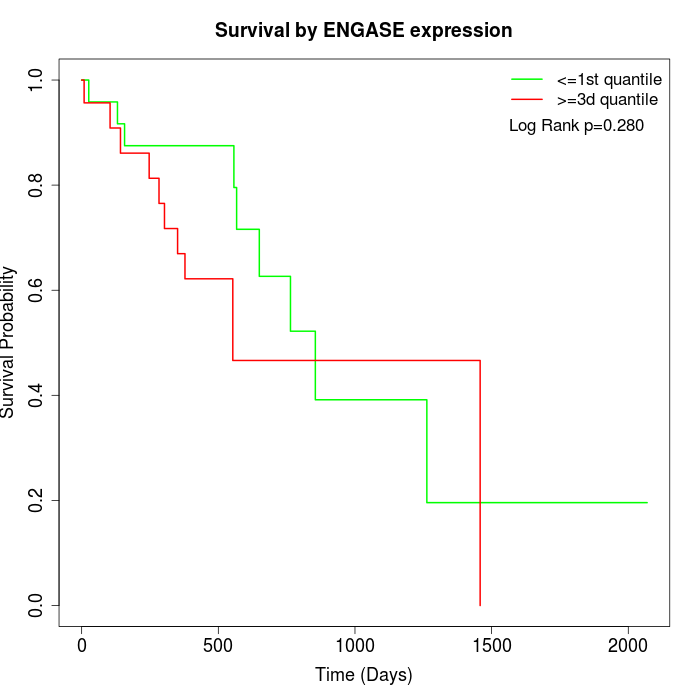
Survival by ENGASE expression:
Note: Click image to view full size file.
Copy number change of ENGASE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ENGASE | 64772 | 4 | 3 | 23 | |
| GSE20123 | ENGASE | 64772 | 4 | 3 | 23 | |
| GSE43470 | ENGASE | 64772 | 3 | 4 | 36 | |
| GSE46452 | ENGASE | 64772 | 33 | 0 | 26 | |
| GSE47630 | ENGASE | 64772 | 8 | 1 | 31 | |
| GSE54993 | ENGASE | 64772 | 2 | 5 | 63 | |
| GSE54994 | ENGASE | 64772 | 10 | 5 | 38 | |
| GSE60625 | ENGASE | 64772 | 6 | 0 | 5 | |
| GSE74703 | ENGASE | 64772 | 3 | 2 | 31 | |
| GSE74704 | ENGASE | 64772 | 4 | 3 | 13 | |
| TCGA | ENGASE | 64772 | 29 | 9 | 58 |
Total number of gains: 106; Total number of losses: 35; Total Number of normals: 347.
Somatic mutations of ENGASE:
Generating mutation plots.
Highly correlated genes for ENGASE:
Showing top 20/256 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ENGASE | KIR3DL3 | 0.775788 | 3 | 0 | 3 |
| ENGASE | GRIPAP1 | 0.765792 | 3 | 0 | 3 |
| ENGASE | LENEP | 0.739971 | 3 | 0 | 3 |
| ENGASE | AQP8 | 0.736891 | 3 | 0 | 3 |
| ENGASE | NXPE4 | 0.733895 | 3 | 0 | 3 |
| ENGASE | OR10H1 | 0.715136 | 4 | 0 | 4 |
| ENGASE | UPK3A | 0.713618 | 3 | 0 | 3 |
| ENGASE | ACSM5 | 0.710593 | 3 | 0 | 3 |
| ENGASE | NOC4L | 0.708247 | 3 | 0 | 3 |
| ENGASE | GRIN2C | 0.706014 | 3 | 0 | 3 |
| ENGASE | ZNF611 | 0.705874 | 3 | 0 | 3 |
| ENGASE | VIPR2 | 0.704782 | 3 | 0 | 3 |
| ENGASE | OR2C1 | 0.68791 | 4 | 0 | 3 |
| ENGASE | SLC1A7 | 0.685849 | 3 | 0 | 3 |
| ENGASE | SMCP | 0.685285 | 4 | 0 | 3 |
| ENGASE | CABP1 | 0.684238 | 3 | 0 | 3 |
| ENGASE | ABCA2 | 0.683318 | 4 | 0 | 3 |
| ENGASE | SPEF1 | 0.683101 | 3 | 0 | 3 |
| ENGASE | CATSPER3 | 0.681679 | 3 | 0 | 3 |
| ENGASE | S1PR4 | 0.679104 | 5 | 0 | 4 |
For details and further investigation, click here