| Full name: epoxide hydrolase 2 | Alias Symbol: ABHD20 | ||
| Type: protein-coding gene | Cytoband: 8p21.2-p21.1 | ||
| Entrez ID: 2053 | HGNC ID: HGNC:3402 | Ensembl Gene: ENSG00000120915 | OMIM ID: 132811 |
| Drug and gene relationship at DGIdb | |||
Expression of EPHX2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPHX2 | 2053 | 209368_at | -1.7288 | 0.0346 | |
| GSE20347 | EPHX2 | 2053 | 209368_at | -1.6433 | 0.0000 | |
| GSE23400 | EPHX2 | 2053 | 209368_at | -0.8317 | 0.0000 | |
| GSE26886 | EPHX2 | 2053 | 209368_at | -2.7220 | 0.0000 | |
| GSE29001 | EPHX2 | 2053 | 209368_at | -1.0403 | 0.0141 | |
| GSE38129 | EPHX2 | 2053 | 209368_at | -1.5998 | 0.0000 | |
| GSE45670 | EPHX2 | 2053 | 209368_at | -1.2643 | 0.0001 | |
| GSE53622 | EPHX2 | 2053 | 72192 | -1.9894 | 0.0000 | |
| GSE53624 | EPHX2 | 2053 | 72192 | -2.1655 | 0.0000 | |
| GSE63941 | EPHX2 | 2053 | 209368_at | 0.2005 | 0.8338 | |
| GSE77861 | EPHX2 | 2053 | 209368_at | -1.5192 | 0.0015 | |
| GSE97050 | EPHX2 | 2053 | A_23_P8834 | -0.7335 | 0.1559 | |
| SRP007169 | EPHX2 | 2053 | RNAseq | -3.1662 | 0.0000 | |
| SRP008496 | EPHX2 | 2053 | RNAseq | -2.7458 | 0.0000 | |
| SRP064894 | EPHX2 | 2053 | RNAseq | -1.9278 | 0.0000 | |
| SRP133303 | EPHX2 | 2053 | RNAseq | -2.4148 | 0.0000 | |
| SRP159526 | EPHX2 | 2053 | RNAseq | -2.3135 | 0.0000 | |
| SRP193095 | EPHX2 | 2053 | RNAseq | -2.1844 | 0.0000 | |
| SRP219564 | EPHX2 | 2053 | RNAseq | -1.9797 | 0.0000 | |
| TCGA | EPHX2 | 2053 | RNAseq | -0.5354 | 0.0029 |
Upregulated datasets: 0; Downregulated datasets: 16.
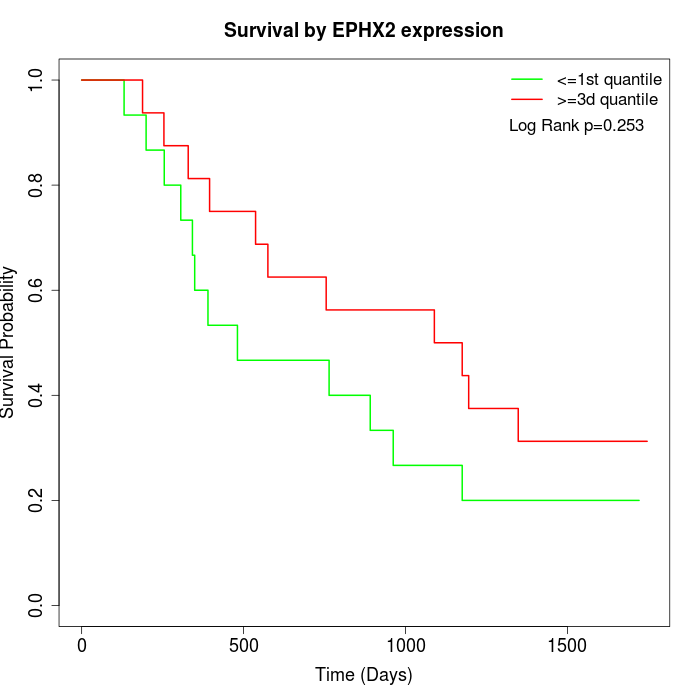
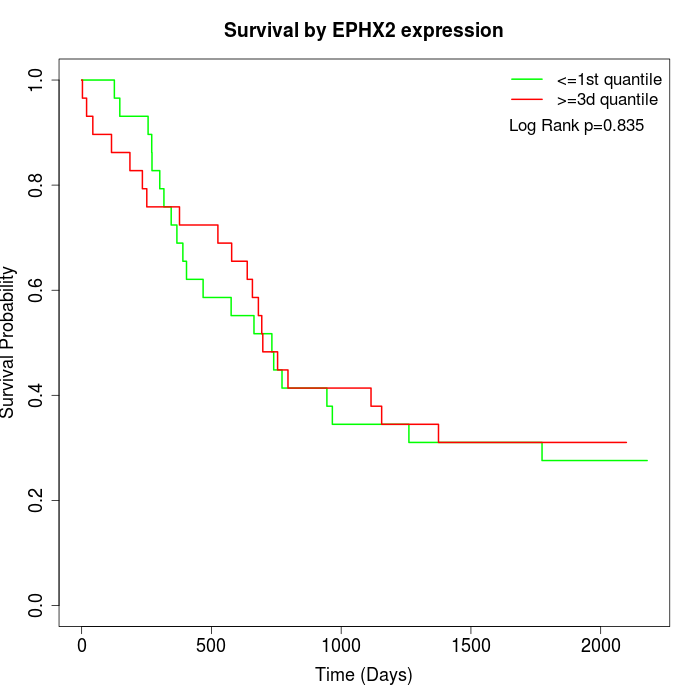
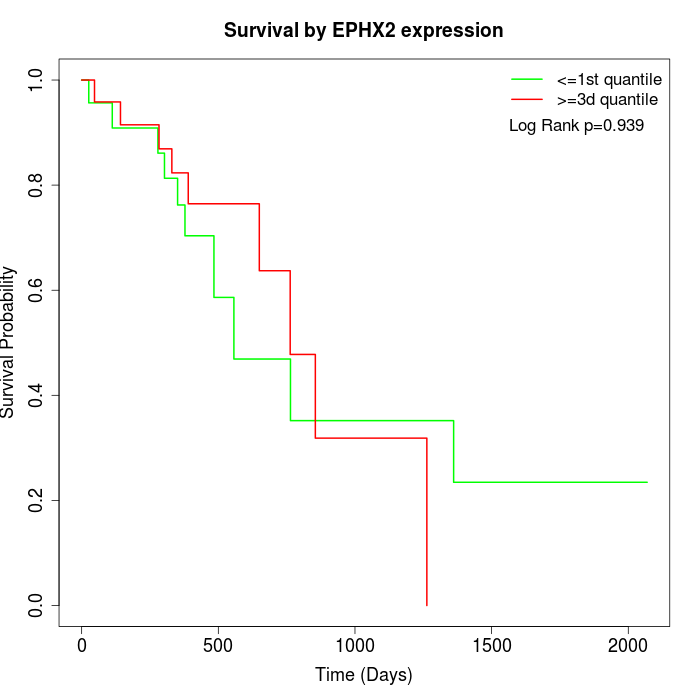
Survival by EPHX2 expression:
Note: Click image to view full size file.
Copy number change of EPHX2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPHX2 | 2053 | 5 | 10 | 15 | |
| GSE20123 | EPHX2 | 2053 | 5 | 10 | 15 | |
| GSE43470 | EPHX2 | 2053 | 4 | 7 | 32 | |
| GSE46452 | EPHX2 | 2053 | 13 | 13 | 33 | |
| GSE47630 | EPHX2 | 2053 | 10 | 8 | 22 | |
| GSE54993 | EPHX2 | 2053 | 2 | 15 | 53 | |
| GSE54994 | EPHX2 | 2053 | 10 | 15 | 28 | |
| GSE60625 | EPHX2 | 2053 | 3 | 0 | 8 | |
| GSE74703 | EPHX2 | 2053 | 4 | 6 | 26 | |
| GSE74704 | EPHX2 | 2053 | 4 | 7 | 9 | |
| TCGA | EPHX2 | 2053 | 16 | 43 | 37 |
Total number of gains: 76; Total number of losses: 134; Total Number of normals: 278.
Somatic mutations of EPHX2:
Generating mutation plots.
Highly correlated genes for EPHX2:
Showing top 20/1296 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPHX2 | CTTNBP2 | 0.834399 | 7 | 0 | 7 |
| EPHX2 | GPD1L | 0.8318 | 10 | 0 | 10 |
| EPHX2 | MYZAP | 0.829072 | 3 | 0 | 3 |
| EPHX2 | SNORA68 | 0.827707 | 4 | 0 | 4 |
| EPHX2 | C5orf66-AS1 | 0.820345 | 6 | 0 | 6 |
| EPHX2 | FCHO2 | 0.813492 | 6 | 0 | 6 |
| EPHX2 | SFTA2 | 0.806225 | 4 | 0 | 4 |
| EPHX2 | MPP7 | 0.803996 | 6 | 0 | 6 |
| EPHX2 | ST6GALNAC1 | 0.799944 | 6 | 0 | 6 |
| EPHX2 | GALNT12 | 0.799504 | 10 | 0 | 10 |
| EPHX2 | NUCB2 | 0.796434 | 10 | 0 | 10 |
| EPHX2 | FAM214A | 0.795456 | 6 | 0 | 6 |
| EPHX2 | PLEKHA7 | 0.794172 | 6 | 0 | 6 |
| EPHX2 | CYSRT1 | 0.790995 | 6 | 0 | 6 |
| EPHX2 | HCG22 | 0.787633 | 6 | 0 | 6 |
| EPHX2 | MTMR10 | 0.7874 | 6 | 0 | 6 |
| EPHX2 | ASPG | 0.787041 | 5 | 0 | 5 |
| EPHX2 | VSIG2 | 0.786534 | 7 | 0 | 7 |
| EPHX2 | RMND5B | 0.786508 | 10 | 0 | 10 |
| EPHX2 | PAIP2B | 0.786116 | 10 | 0 | 10 |
For details and further investigation, click here