| Full name: surfactant associated 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 389376 | HGNC ID: HGNC:18386 | Ensembl Gene: ENSG00000196260 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of SFTA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SFTA2 | 389376 | 244056_at | -4.0434 | 0.0400 | |
| GSE26886 | SFTA2 | 389376 | 244056_at | -3.6791 | 0.0000 | |
| GSE45670 | SFTA2 | 389376 | 244056_at | -2.8424 | 0.0028 | |
| GSE63941 | SFTA2 | 389376 | 244056_at | 0.2265 | 0.6391 | |
| GSE77861 | SFTA2 | 389376 | 244056_at | -3.6585 | 0.0003 | |
| SRP007169 | SFTA2 | 389376 | RNAseq | -12.3242 | 0.0000 | |
| SRP008496 | SFTA2 | 389376 | RNAseq | -8.1666 | 0.0000 | |
| SRP064894 | SFTA2 | 389376 | RNAseq | -6.7097 | 0.0000 | |
| SRP133303 | SFTA2 | 389376 | RNAseq | -8.6637 | 0.0000 | |
| SRP159526 | SFTA2 | 389376 | RNAseq | -8.4820 | 0.0000 | |
| TCGA | SFTA2 | 389376 | RNAseq | -2.0654 | 0.0910 |
Upregulated datasets: 0; Downregulated datasets: 9.
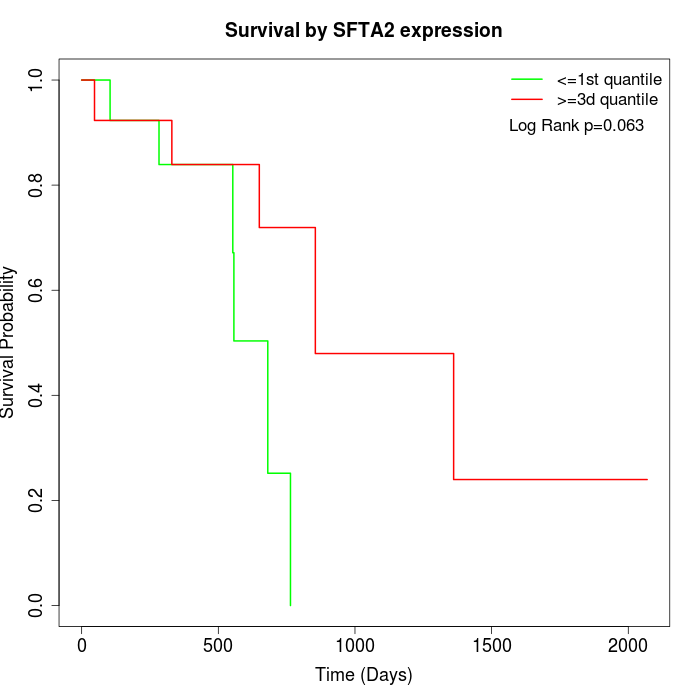
Survival by SFTA2 expression:
Note: Click image to view full size file.
Copy number change of SFTA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SFTA2 | 389376 | 5 | 1 | 24 | |
| GSE20123 | SFTA2 | 389376 | 5 | 1 | 24 | |
| GSE43470 | SFTA2 | 389376 | 6 | 1 | 36 | |
| GSE46452 | SFTA2 | 389376 | 1 | 10 | 48 | |
| GSE47630 | SFTA2 | 389376 | 7 | 3 | 30 | |
| GSE54993 | SFTA2 | 389376 | 2 | 1 | 67 | |
| GSE54994 | SFTA2 | 389376 | 11 | 4 | 38 | |
| GSE60625 | SFTA2 | 389376 | 0 | 1 | 10 | |
| GSE74703 | SFTA2 | 389376 | 6 | 0 | 30 | |
| GSE74704 | SFTA2 | 389376 | 2 | 0 | 18 | |
| TCGA | SFTA2 | 389376 | 17 | 16 | 63 |
Total number of gains: 62; Total number of losses: 38; Total Number of normals: 388.
Somatic mutations of SFTA2:
Generating mutation plots.
Highly correlated genes for SFTA2:
Showing top 20/1237 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SFTA2 | GYS2 | 0.930062 | 4 | 0 | 4 |
| SFTA2 | SLC16A6 | 0.899407 | 4 | 0 | 4 |
| SFTA2 | TP53I3 | 0.897938 | 4 | 0 | 4 |
| SFTA2 | PADI1 | 0.896614 | 4 | 0 | 4 |
| SFTA2 | RAET1E | 0.893266 | 4 | 0 | 4 |
| SFTA2 | TMEM40 | 0.889684 | 4 | 0 | 4 |
| SFTA2 | RANBP9 | 0.888665 | 4 | 0 | 4 |
| SFTA2 | PSCA | 0.883194 | 4 | 0 | 4 |
| SFTA2 | ENDOU | 0.883114 | 4 | 0 | 4 |
| SFTA2 | EPB41L3 | 0.882391 | 4 | 0 | 4 |
| SFTA2 | C5orf66-AS1 | 0.881741 | 4 | 0 | 4 |
| SFTA2 | UBL3 | 0.879564 | 4 | 0 | 4 |
| SFTA2 | NUCB2 | 0.87936 | 4 | 0 | 4 |
| SFTA2 | KLB | 0.878666 | 4 | 0 | 4 |
| SFTA2 | EPS8L2 | 0.876922 | 4 | 0 | 4 |
| SFTA2 | ESPL1 | 0.870871 | 4 | 0 | 4 |
| SFTA2 | VAT1 | 0.870493 | 4 | 0 | 4 |
| SFTA2 | CAPN5 | 0.870169 | 4 | 0 | 4 |
| SFTA2 | FLG | 0.867145 | 4 | 0 | 4 |
| SFTA2 | ARHGAP27 | 0.866961 | 4 | 0 | 4 |
For details and further investigation, click here