| Full name: epoxide hydrolase 4 | Alias Symbol: EPHXRP|FLJ90341|EH4 | ||
| Type: protein-coding gene | Cytoband: 1p22.1 | ||
| Entrez ID: 253152 | HGNC ID: HGNC:23758 | Ensembl Gene: ENSG00000172031 | OMIM ID: 617401 |
| Drug and gene relationship at DGIdb | |||
Expression of EPHX4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPHX4 | 253152 | 239579_at | -0.0827 | 0.8155 | |
| GSE26886 | EPHX4 | 253152 | 239579_at | 0.5395 | 0.0291 | |
| GSE45670 | EPHX4 | 253152 | 239579_at | -0.2701 | 0.1020 | |
| GSE53622 | EPHX4 | 253152 | 31090 | 0.1170 | 0.5965 | |
| GSE53624 | EPHX4 | 253152 | 31090 | 0.4956 | 0.0098 | |
| GSE63941 | EPHX4 | 253152 | 239579_at | -1.3557 | 0.2796 | |
| GSE77861 | EPHX4 | 253152 | 239579_at | 0.0441 | 0.8126 | |
| SRP159526 | EPHX4 | 253152 | RNAseq | 1.2900 | 0.1248 | |
| TCGA | EPHX4 | 253152 | RNAseq | 0.5408 | 0.2472 |
Upregulated datasets: 0; Downregulated datasets: 0.
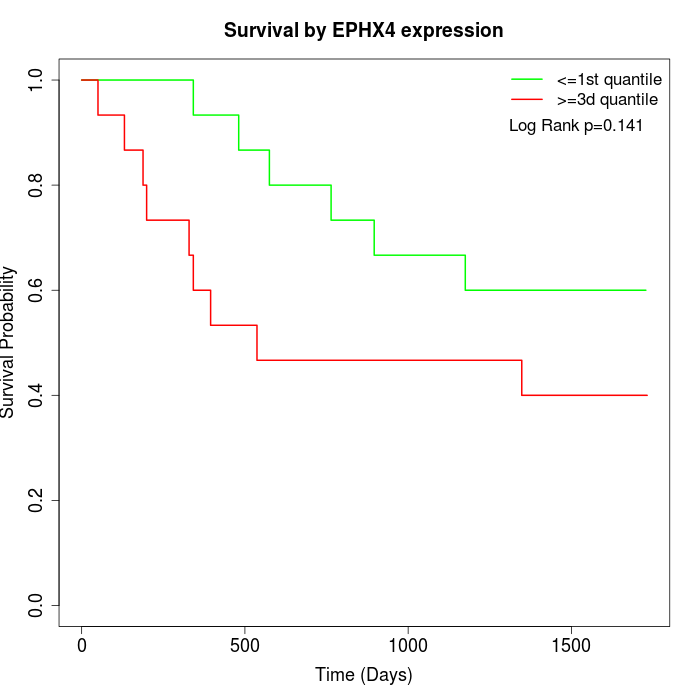
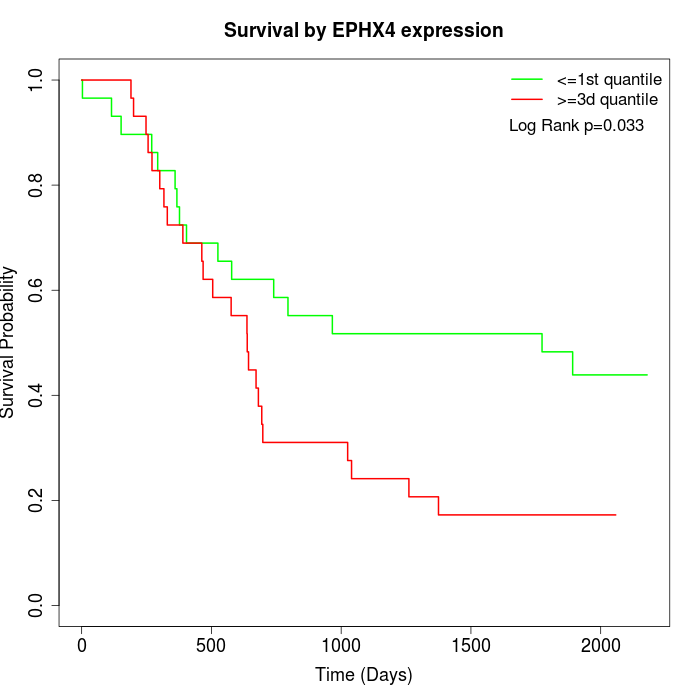
Survival by EPHX4 expression:
Note: Click image to view full size file.
Copy number change of EPHX4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPHX4 | 253152 | 0 | 8 | 22 | |
| GSE20123 | EPHX4 | 253152 | 0 | 8 | 22 | |
| GSE43470 | EPHX4 | 253152 | 2 | 5 | 36 | |
| GSE46452 | EPHX4 | 253152 | 1 | 1 | 57 | |
| GSE47630 | EPHX4 | 253152 | 8 | 5 | 27 | |
| GSE54993 | EPHX4 | 253152 | 0 | 1 | 69 | |
| GSE54994 | EPHX4 | 253152 | 6 | 3 | 44 | |
| GSE60625 | EPHX4 | 253152 | 0 | 0 | 11 | |
| GSE74703 | EPHX4 | 253152 | 1 | 4 | 31 | |
| GSE74704 | EPHX4 | 253152 | 0 | 5 | 15 | |
| TCGA | EPHX4 | 253152 | 6 | 24 | 66 |
Total number of gains: 24; Total number of losses: 64; Total Number of normals: 400.
Somatic mutations of EPHX4:
Generating mutation plots.
Highly correlated genes for EPHX4:
Showing top 20/110 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPHX4 | CLDN12 | 0.726426 | 3 | 0 | 3 |
| EPHX4 | SRBD1 | 0.665749 | 3 | 0 | 3 |
| EPHX4 | KCNE3 | 0.657616 | 3 | 0 | 3 |
| EPHX4 | NHS | 0.646361 | 3 | 0 | 3 |
| EPHX4 | SNRNP25 | 0.638316 | 3 | 0 | 3 |
| EPHX4 | FBXO4 | 0.628621 | 4 | 0 | 3 |
| EPHX4 | ARL16 | 0.625671 | 3 | 0 | 3 |
| EPHX4 | EDRF1 | 0.61373 | 3 | 0 | 3 |
| EPHX4 | MTFMT | 0.613596 | 3 | 0 | 3 |
| EPHX4 | ADSL | 0.61287 | 3 | 0 | 3 |
| EPHX4 | MID1 | 0.612113 | 3 | 0 | 3 |
| EPHX4 | IFT46 | 0.60881 | 3 | 0 | 3 |
| EPHX4 | ZNF599 | 0.607432 | 4 | 0 | 3 |
| EPHX4 | EMILIN2 | 0.602718 | 3 | 0 | 3 |
| EPHX4 | CLSTN2 | 0.601718 | 4 | 0 | 4 |
| EPHX4 | DPY19L2 | 0.600861 | 4 | 0 | 3 |
| EPHX4 | FAM171B | 0.595009 | 5 | 0 | 3 |
| EPHX4 | F13A1 | 0.589645 | 3 | 0 | 3 |
| EPHX4 | HNRNPLL | 0.586379 | 5 | 0 | 4 |
| EPHX4 | PLCL1 | 0.585686 | 4 | 0 | 3 |
For details and further investigation, click here