| Full name: midline 1 | Alias Symbol: OS|FXY|TRIM18|RNF59 | ||
| Type: protein-coding gene | Cytoband: Xp22 | ||
| Entrez ID: 4281 | HGNC ID: HGNC:7095 | Ensembl Gene: ENSG00000101871 | OMIM ID: 300552 |
| Drug and gene relationship at DGIdb | |||
Expression of MID1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MID1 | 4281 | 203636_at | -0.1488 | 0.8404 | |
| GSE20347 | MID1 | 4281 | 203637_s_at | -0.4507 | 0.2782 | |
| GSE23400 | MID1 | 4281 | 203636_at | -0.0477 | 0.5274 | |
| GSE26886 | MID1 | 4281 | 203637_s_at | 0.3596 | 0.2144 | |
| GSE29001 | MID1 | 4281 | 203637_s_at | -0.0778 | 0.8450 | |
| GSE38129 | MID1 | 4281 | 203636_at | -0.5614 | 0.0108 | |
| GSE45670 | MID1 | 4281 | 203636_at | 0.1302 | 0.6451 | |
| GSE53622 | MID1 | 4281 | 42393 | 0.0828 | 0.4070 | |
| GSE53624 | MID1 | 4281 | 42393 | -0.0059 | 0.9513 | |
| GSE63941 | MID1 | 4281 | 203637_s_at | -0.2073 | 0.8158 | |
| GSE77861 | MID1 | 4281 | 203637_s_at | 0.1675 | 0.6523 | |
| GSE97050 | MID1 | A_21_P0000116 | 0.1657 | 0.5478 | ||
| SRP007169 | MID1 | 4281 | RNAseq | 0.5020 | 0.3332 | |
| SRP008496 | MID1 | 4281 | RNAseq | 1.0816 | 0.0149 | |
| SRP064894 | MID1 | 4281 | RNAseq | 0.0339 | 0.8996 | |
| SRP133303 | MID1 | 4281 | RNAseq | 0.9179 | 0.0500 | |
| SRP159526 | MID1 | 4281 | RNAseq | 0.0640 | 0.7777 | |
| SRP193095 | MID1 | 4281 | RNAseq | 0.2271 | 0.2161 | |
| SRP219564 | MID1 | 4281 | RNAseq | 0.2270 | 0.6015 | |
| TCGA | MID1 | 4281 | RNAseq | 0.0115 | 0.8840 |
Upregulated datasets: 1; Downregulated datasets: 0.
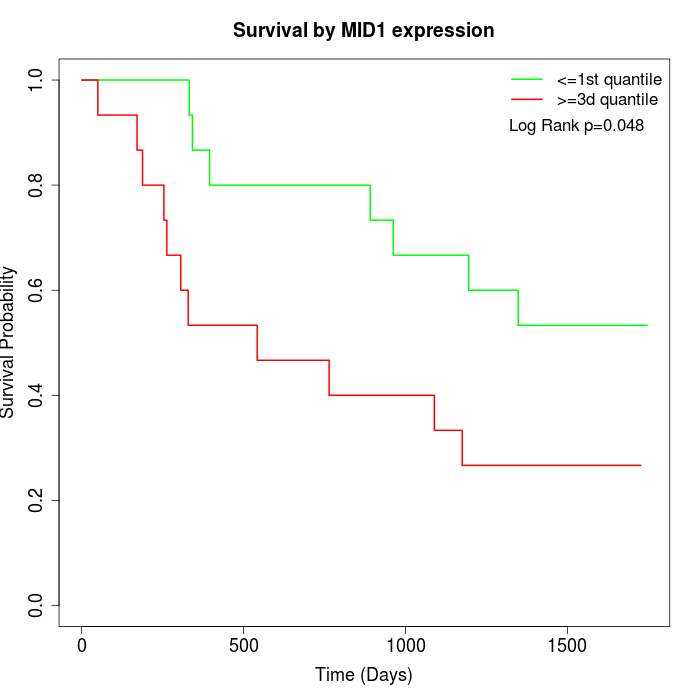
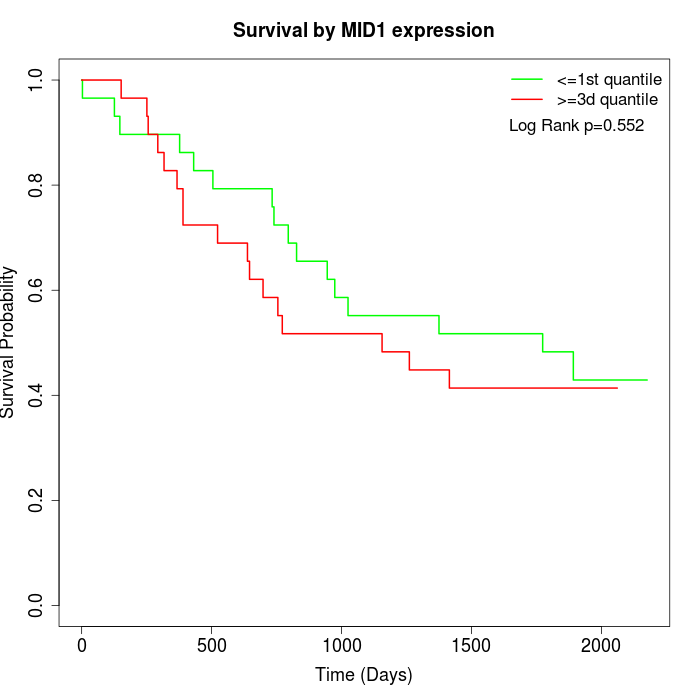
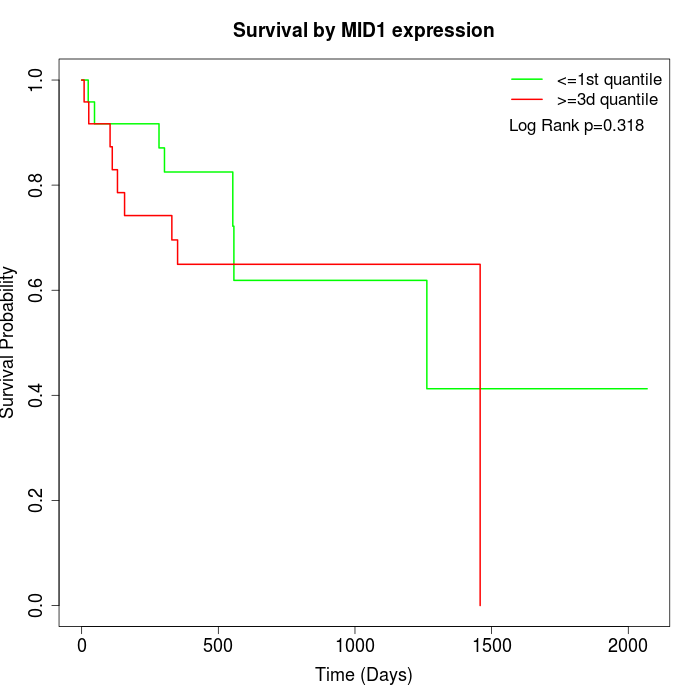
Survival by MID1 expression:
Note: Click image to view full size file.
Copy number change of MID1:
No record found for this gene.
Somatic mutations of MID1:
Generating mutation plots.
Highly correlated genes for MID1:
Showing top 20/33 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MID1 | CLEC12B | 0.781703 | 3 | 0 | 3 |
| MID1 | AKR7A3 | 0.713751 | 3 | 0 | 3 |
| MID1 | HS2ST1 | 0.709985 | 3 | 0 | 3 |
| MID1 | WNT2B | 0.704495 | 3 | 0 | 3 |
| MID1 | ZNF682 | 0.696027 | 3 | 0 | 3 |
| MID1 | RASSF6 | 0.674789 | 4 | 0 | 3 |
| MID1 | ZNF500 | 0.672923 | 3 | 0 | 3 |
| MID1 | HOGA1 | 0.670565 | 3 | 0 | 3 |
| MID1 | DDX43 | 0.667329 | 3 | 0 | 3 |
| MID1 | TRIM2 | 0.657983 | 4 | 0 | 3 |
| MID1 | RSPH4A | 0.64962 | 3 | 0 | 3 |
| MID1 | TMEM243 | 0.637531 | 3 | 0 | 3 |
| MID1 | ABAT | 0.617306 | 4 | 0 | 3 |
| MID1 | TNFSF13B | 0.616731 | 3 | 0 | 3 |
| MID1 | TBC1D9 | 0.615637 | 4 | 0 | 3 |
| MID1 | EPHX4 | 0.612113 | 3 | 0 | 3 |
| MID1 | COQ5 | 0.611208 | 4 | 0 | 3 |
| MID1 | CCDC113 | 0.607838 | 3 | 0 | 3 |
| MID1 | KAT6A | 0.604656 | 4 | 0 | 3 |
| MID1 | GEMIN8 | 0.599262 | 5 | 0 | 3 |
For details and further investigation, click here