| Full name: phospholipase C like 1 (inactive) | Alias Symbol: PLC-L|PLCL|PRIP|PPP1R127 | ||
| Type: protein-coding gene | Cytoband: 2q33.1 | ||
| Entrez ID: 5334 | HGNC ID: HGNC:9063 | Ensembl Gene: ENSG00000115896 | OMIM ID: 600597 |
| Drug and gene relationship at DGIdb | |||
Expression of PLCL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLCL1 | 5334 | 205934_at | -0.5300 | 0.7944 | |
| GSE20347 | PLCL1 | 5334 | 205934_at | 0.0342 | 0.9264 | |
| GSE23400 | PLCL1 | 5334 | 205934_at | -0.0910 | 0.3479 | |
| GSE26886 | PLCL1 | 5334 | 205934_at | 0.6195 | 0.0570 | |
| GSE29001 | PLCL1 | 5334 | 205934_at | 0.0682 | 0.7854 | |
| GSE38129 | PLCL1 | 5334 | 205934_at | -0.4448 | 0.2869 | |
| GSE45670 | PLCL1 | 5334 | 205934_at | -2.0127 | 0.0000 | |
| GSE53622 | PLCL1 | 5334 | 46448 | -0.9177 | 0.0002 | |
| GSE53624 | PLCL1 | 5334 | 46448 | -0.3502 | 0.0876 | |
| GSE63941 | PLCL1 | 5334 | 205934_at | -1.5676 | 0.0000 | |
| GSE77861 | PLCL1 | 5334 | 241859_at | -0.1032 | 0.2952 | |
| GSE97050 | PLCL1 | 5334 | A_33_P3289865 | -0.9199 | 0.1623 | |
| SRP007169 | PLCL1 | 5334 | RNAseq | 2.2197 | 0.0210 | |
| SRP064894 | PLCL1 | 5334 | RNAseq | 0.0559 | 0.8823 | |
| SRP133303 | PLCL1 | 5334 | RNAseq | -0.0184 | 0.9655 | |
| SRP159526 | PLCL1 | 5334 | RNAseq | 0.5710 | 0.2880 | |
| SRP193095 | PLCL1 | 5334 | RNAseq | 0.5912 | 0.0193 | |
| SRP219564 | PLCL1 | 5334 | RNAseq | -0.7858 | 0.4483 | |
| TCGA | PLCL1 | 5334 | RNAseq | -0.8584 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 2.
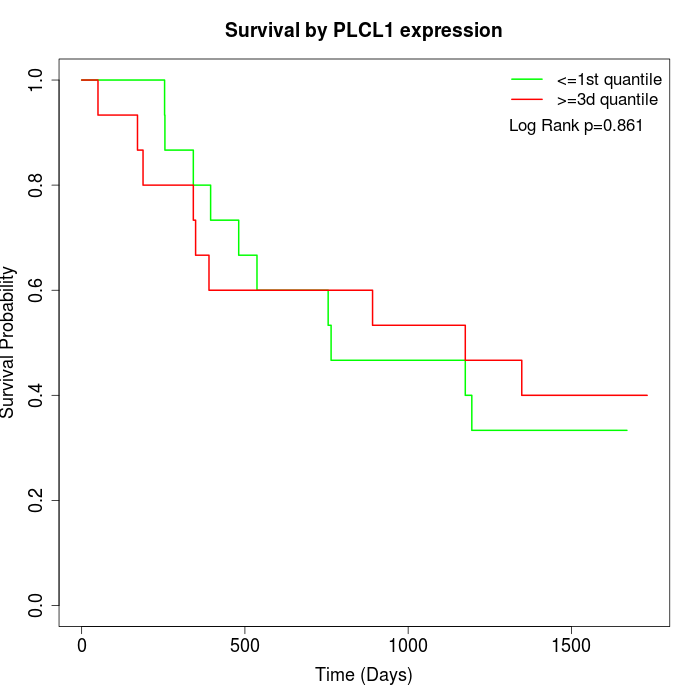
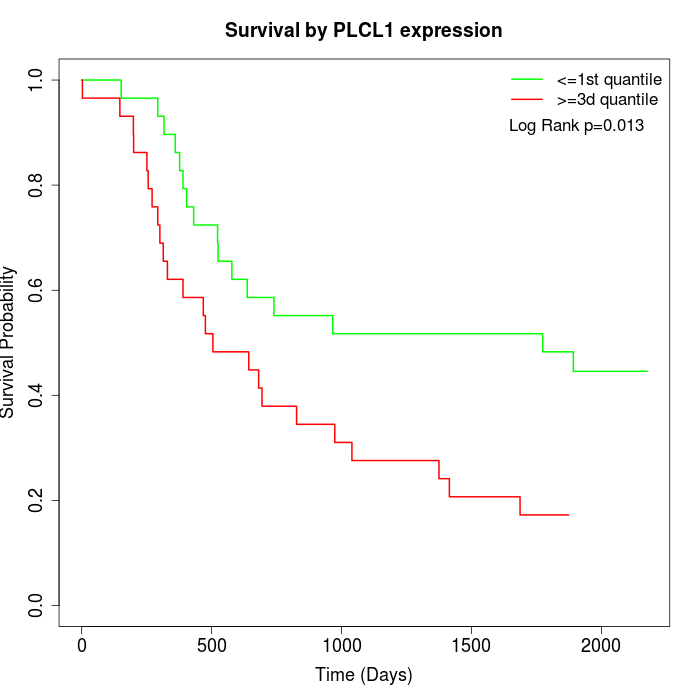
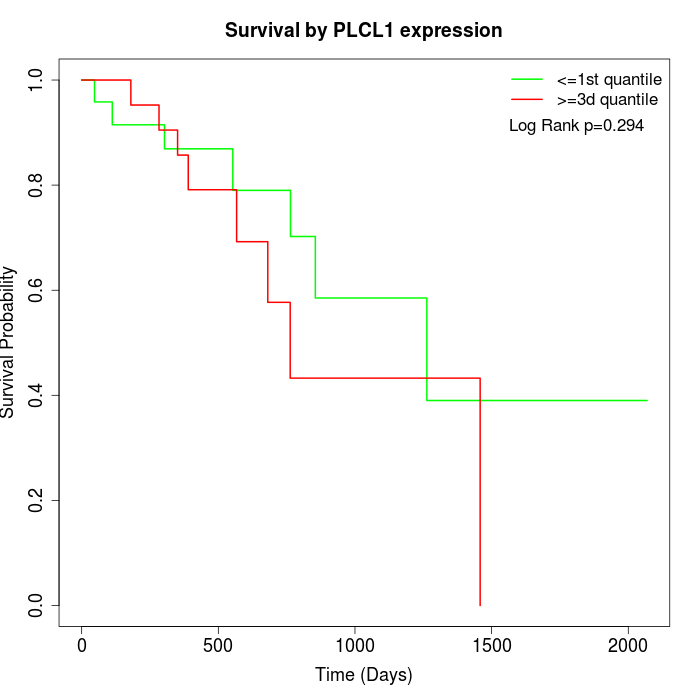
Survival by PLCL1 expression:
Note: Click image to view full size file.
Copy number change of PLCL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLCL1 | 5334 | 9 | 3 | 18 | |
| GSE20123 | PLCL1 | 5334 | 9 | 3 | 18 | |
| GSE43470 | PLCL1 | 5334 | 3 | 1 | 39 | |
| GSE46452 | PLCL1 | 5334 | 1 | 4 | 54 | |
| GSE47630 | PLCL1 | 5334 | 4 | 5 | 31 | |
| GSE54993 | PLCL1 | 5334 | 0 | 3 | 67 | |
| GSE54994 | PLCL1 | 5334 | 10 | 8 | 35 | |
| GSE60625 | PLCL1 | 5334 | 0 | 3 | 8 | |
| GSE74703 | PLCL1 | 5334 | 2 | 1 | 33 | |
| GSE74704 | PLCL1 | 5334 | 4 | 3 | 13 | |
| TCGA | PLCL1 | 5334 | 23 | 10 | 63 |
Total number of gains: 65; Total number of losses: 44; Total Number of normals: 379.
Somatic mutations of PLCL1:
Generating mutation plots.
Highly correlated genes for PLCL1:
Showing top 20/1126 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLCL1 | TNFSF12 | 0.804063 | 3 | 0 | 3 |
| PLCL1 | TAB1 | 0.771308 | 3 | 0 | 3 |
| PLCL1 | EPHA3 | 0.770601 | 7 | 0 | 7 |
| PLCL1 | DIRAS1 | 0.769223 | 3 | 0 | 3 |
| PLCL1 | FIBIN | 0.76887 | 4 | 0 | 4 |
| PLCL1 | FRMD3 | 0.765107 | 5 | 0 | 5 |
| PLCL1 | RASL12 | 0.757168 | 10 | 0 | 9 |
| PLCL1 | PGR | 0.752182 | 6 | 0 | 6 |
| PLCL1 | TMEM170B | 0.748713 | 6 | 0 | 6 |
| PLCL1 | EPHA7 | 0.748589 | 6 | 0 | 6 |
| PLCL1 | FAM13C | 0.747784 | 4 | 0 | 3 |
| PLCL1 | MECP2 | 0.745945 | 4 | 0 | 4 |
| PLCL1 | LRCH2 | 0.741811 | 7 | 0 | 6 |
| PLCL1 | KCNT2 | 0.741042 | 7 | 0 | 7 |
| PLCL1 | TBX5 | 0.733594 | 7 | 0 | 7 |
| PLCL1 | TPPP | 0.733339 | 3 | 0 | 3 |
| PLCL1 | RBPMS2 | 0.731804 | 6 | 0 | 6 |
| PLCL1 | DIXDC1 | 0.730569 | 10 | 0 | 9 |
| PLCL1 | XKR4 | 0.730425 | 5 | 0 | 4 |
| PLCL1 | PCMTD1 | 0.729644 | 4 | 0 | 4 |
For details and further investigation, click here