| Full name: family with sequence similarity 199, X-linked | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xq22.2 | ||
| Entrez ID: 139231 | HGNC ID: HGNC:25195 | Ensembl Gene: ENSG00000123575 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of FAM199X:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAM199X | 139231 | 225216_at | -0.0862 | 0.8746 | |
| GSE26886 | FAM199X | 139231 | 227133_at | -0.3993 | 0.1929 | |
| GSE45670 | FAM199X | 139231 | 227133_at | 0.5835 | 0.0027 | |
| GSE53622 | FAM199X | 139231 | 68167 | 0.1194 | 0.0632 | |
| GSE53624 | FAM199X | 139231 | 68167 | 0.1148 | 0.0332 | |
| GSE63941 | FAM199X | 139231 | 227133_at | 1.2414 | 0.0007 | |
| GSE77861 | FAM199X | 139231 | 225216_at | 0.2393 | 0.2463 | |
| GSE97050 | FAM199X | 139231 | A_23_P45294 | -0.1314 | 0.7329 | |
| SRP007169 | FAM199X | 139231 | RNAseq | 0.0181 | 0.9648 | |
| SRP008496 | FAM199X | 139231 | RNAseq | 0.3361 | 0.1509 | |
| SRP064894 | FAM199X | 139231 | RNAseq | -0.0070 | 0.9748 | |
| SRP133303 | FAM199X | 139231 | RNAseq | 0.4920 | 0.0385 | |
| SRP159526 | FAM199X | 139231 | RNAseq | -0.1194 | 0.5488 | |
| SRP193095 | FAM199X | 139231 | RNAseq | -0.0965 | 0.3147 | |
| SRP219564 | FAM199X | 139231 | RNAseq | -0.0349 | 0.9141 | |
| TCGA | FAM199X | 139231 | RNAseq | 0.0110 | 0.8365 |
Upregulated datasets: 1; Downregulated datasets: 0.
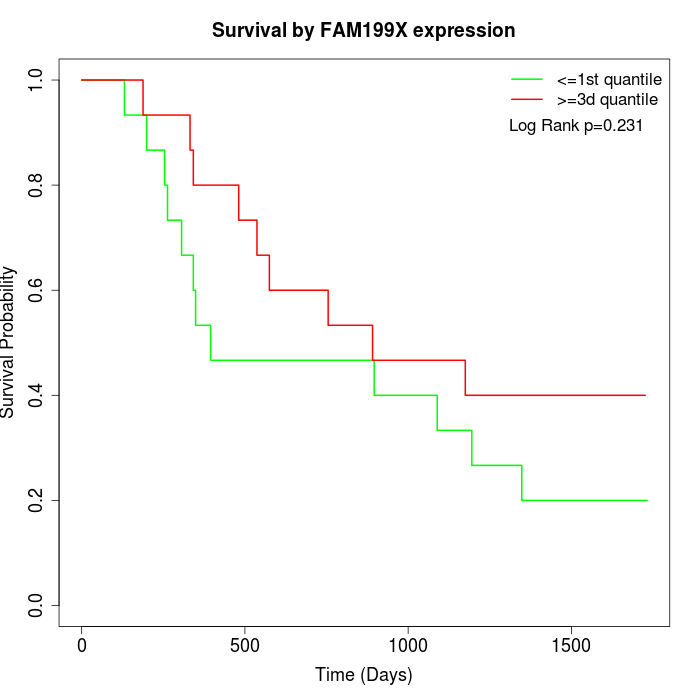
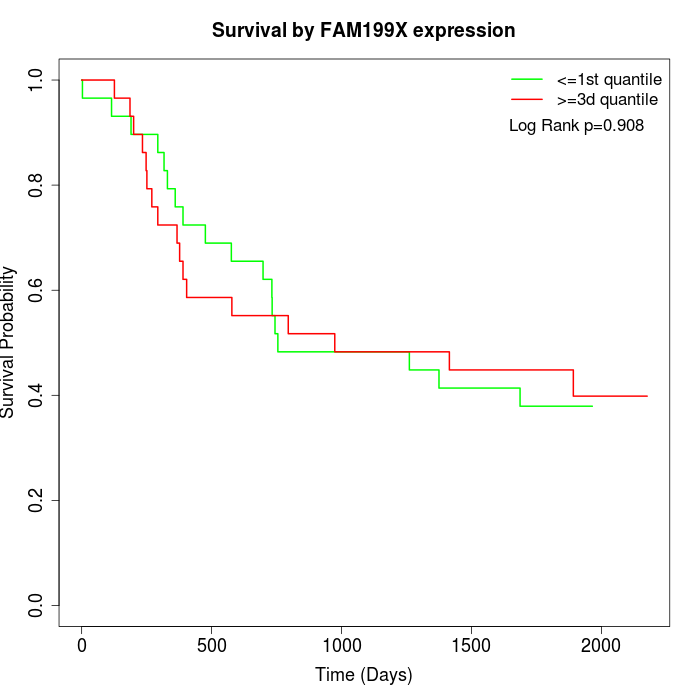
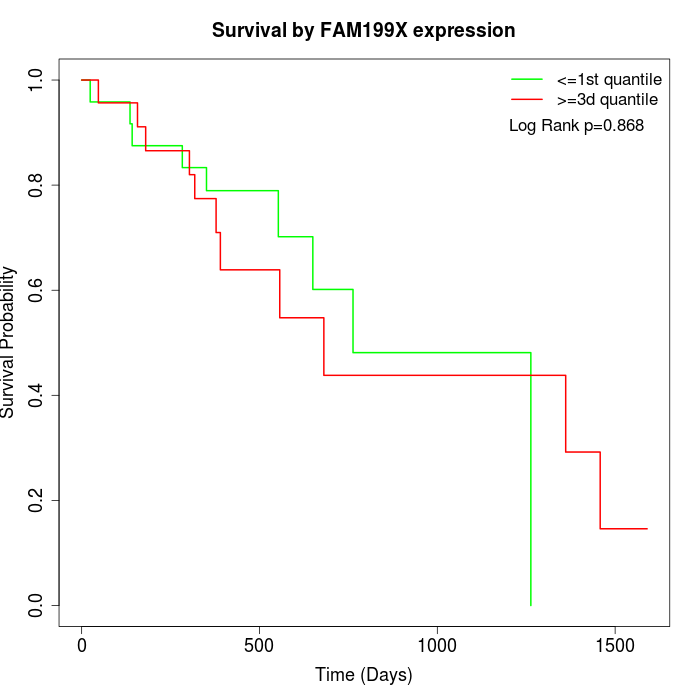
Survival by FAM199X expression:
Note: Click image to view full size file.
Copy number change of FAM199X:
No record found for this gene.
Somatic mutations of FAM199X:
Generating mutation plots.
Highly correlated genes for FAM199X:
Showing top 20/172 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAM199X | PDE6D | 0.822317 | 3 | 0 | 3 |
| FAM199X | LMBR1 | 0.782607 | 3 | 0 | 3 |
| FAM199X | PIGA | 0.762982 | 3 | 0 | 3 |
| FAM199X | FAM76B | 0.761946 | 3 | 0 | 3 |
| FAM199X | KLHL11 | 0.758865 | 3 | 0 | 3 |
| FAM199X | TPMT | 0.757009 | 3 | 0 | 3 |
| FAM199X | AGBL5 | 0.754327 | 3 | 0 | 3 |
| FAM199X | ATG4C | 0.744747 | 3 | 0 | 3 |
| FAM199X | MCM3 | 0.73653 | 3 | 0 | 3 |
| FAM199X | ZXDB | 0.736248 | 3 | 0 | 3 |
| FAM199X | NKAP | 0.736208 | 3 | 0 | 3 |
| FAM199X | FAM168A | 0.734017 | 3 | 0 | 3 |
| FAM199X | MRPL38 | 0.732953 | 3 | 0 | 3 |
| FAM199X | ANKRD40 | 0.732749 | 3 | 0 | 3 |
| FAM199X | SPRED2 | 0.730967 | 3 | 0 | 3 |
| FAM199X | NCBP1 | 0.73 | 3 | 0 | 3 |
| FAM199X | FCHSD2 | 0.72954 | 3 | 0 | 3 |
| FAM199X | C17orf80 | 0.728841 | 3 | 0 | 3 |
| FAM199X | ZFX | 0.728016 | 3 | 0 | 3 |
| FAM199X | RAC1 | 0.723721 | 3 | 0 | 3 |
For details and further investigation, click here