| Full name: growth hormone inducible transmembrane protein | Alias Symbol: HSPC282|PTD010|DERP2|My021|TMBIM5 | ||
| Type: protein-coding gene | Cytoband: 10q23.1 | ||
| Entrez ID: 27069 | HGNC ID: HGNC:17281 | Ensembl Gene: ENSG00000165678 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of GHITM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GHITM | 27069 | 209249_s_at | -0.3257 | 0.3035 | |
| GSE20347 | GHITM | 27069 | 209249_s_at | -0.5050 | 0.0015 | |
| GSE23400 | GHITM | 27069 | 209249_s_at | -0.1919 | 0.0075 | |
| GSE26886 | GHITM | 27069 | 209249_s_at | -1.3293 | 0.0000 | |
| GSE29001 | GHITM | 27069 | 209249_s_at | -0.5498 | 0.0706 | |
| GSE38129 | GHITM | 27069 | 209249_s_at | -0.2230 | 0.1176 | |
| GSE45670 | GHITM | 27069 | 209249_s_at | -0.0880 | 0.5096 | |
| GSE53622 | GHITM | 27069 | 114424 | -0.5094 | 0.0000 | |
| GSE53624 | GHITM | 27069 | 114424 | -0.3403 | 0.0000 | |
| GSE63941 | GHITM | 27069 | 209249_s_at | 0.5137 | 0.1400 | |
| GSE77861 | GHITM | 27069 | 209249_s_at | -0.4093 | 0.1380 | |
| GSE97050 | GHITM | 27069 | A_33_P3217073 | -0.4460 | 0.1466 | |
| SRP007169 | GHITM | 27069 | RNAseq | -0.9743 | 0.0420 | |
| SRP008496 | GHITM | 27069 | RNAseq | -0.8941 | 0.0010 | |
| SRP064894 | GHITM | 27069 | RNAseq | -0.6312 | 0.0000 | |
| SRP133303 | GHITM | 27069 | RNAseq | -0.0006 | 0.9964 | |
| SRP159526 | GHITM | 27069 | RNAseq | -0.0074 | 0.9886 | |
| SRP193095 | GHITM | 27069 | RNAseq | -0.5698 | 0.0000 | |
| SRP219564 | GHITM | 27069 | RNAseq | -0.3959 | 0.0453 | |
| TCGA | GHITM | 27069 | RNAseq | -0.1400 | 0.0008 |
Upregulated datasets: 0; Downregulated datasets: 1.
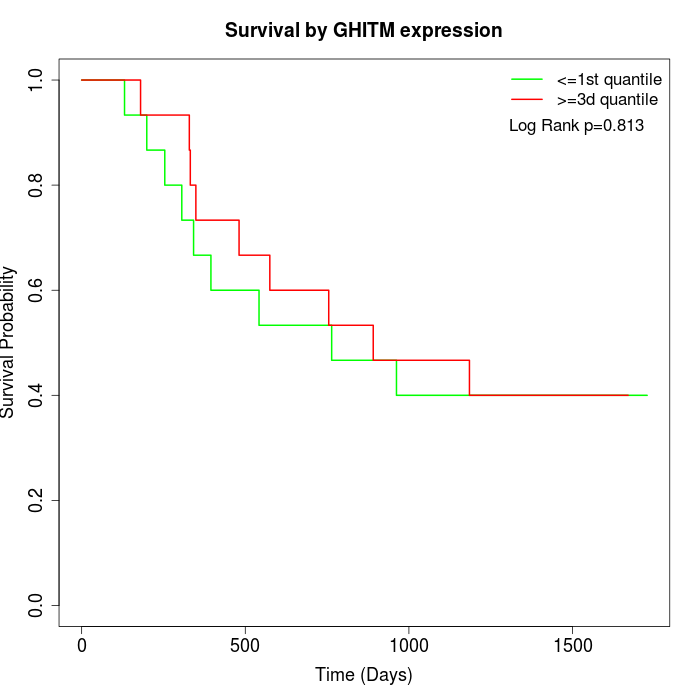
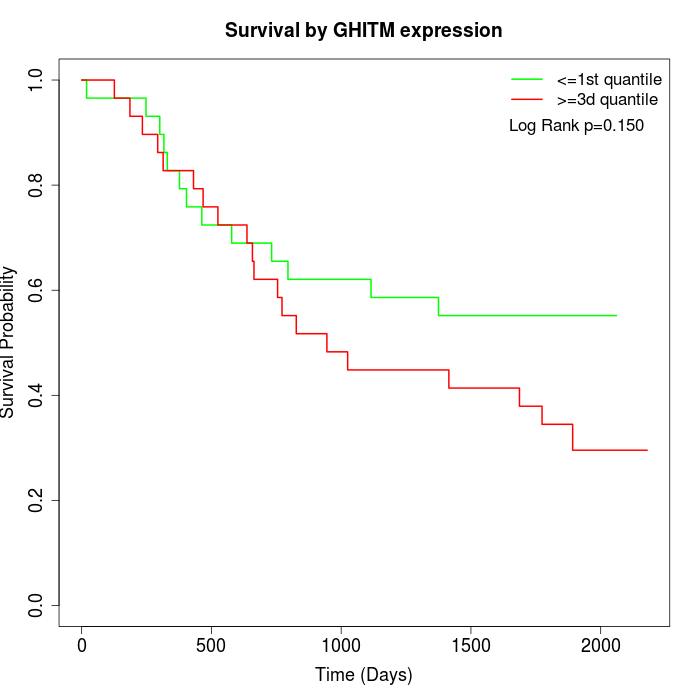
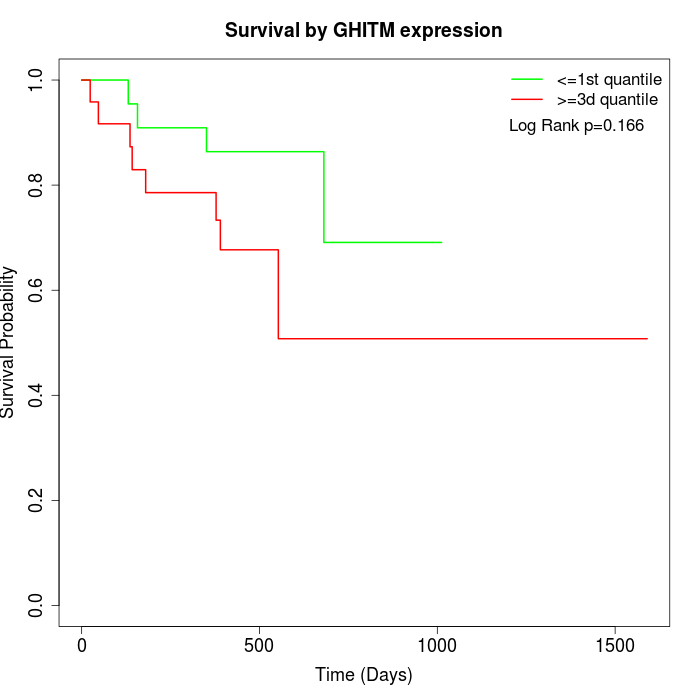
Survival by GHITM expression:
Note: Click image to view full size file.
Copy number change of GHITM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GHITM | 27069 | 1 | 6 | 23 | |
| GSE20123 | GHITM | 27069 | 1 | 5 | 24 | |
| GSE43470 | GHITM | 27069 | 1 | 6 | 36 | |
| GSE46452 | GHITM | 27069 | 0 | 11 | 48 | |
| GSE47630 | GHITM | 27069 | 2 | 14 | 24 | |
| GSE54993 | GHITM | 27069 | 7 | 0 | 63 | |
| GSE54994 | GHITM | 27069 | 1 | 12 | 40 | |
| GSE60625 | GHITM | 27069 | 0 | 0 | 11 | |
| GSE74703 | GHITM | 27069 | 1 | 4 | 31 | |
| GSE74704 | GHITM | 27069 | 0 | 3 | 17 | |
| TCGA | GHITM | 27069 | 9 | 25 | 62 |
Total number of gains: 23; Total number of losses: 86; Total Number of normals: 379.
Somatic mutations of GHITM:
Generating mutation plots.
Highly correlated genes for GHITM:
Showing top 20/991 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GHITM | ESYT2 | 0.768732 | 4 | 0 | 4 |
| GHITM | PRPF38A | 0.758214 | 4 | 0 | 4 |
| GHITM | CCDC25 | 0.748804 | 3 | 0 | 3 |
| GHITM | IMPACT | 0.743488 | 3 | 0 | 3 |
| GHITM | NTAN1 | 0.731394 | 3 | 0 | 3 |
| GHITM | TBC1D20 | 0.729507 | 3 | 0 | 3 |
| GHITM | KIF13B | 0.727103 | 4 | 0 | 4 |
| GHITM | PPP3CC | 0.725385 | 3 | 0 | 3 |
| GHITM | PSEN2 | 0.714845 | 3 | 0 | 3 |
| GHITM | LMO4 | 0.713055 | 3 | 0 | 3 |
| GHITM | ANAPC16 | 0.707312 | 4 | 0 | 4 |
| GHITM | CBY1 | 0.707275 | 3 | 0 | 3 |
| GHITM | MPP7 | 0.70654 | 7 | 0 | 6 |
| GHITM | NPTN | 0.700818 | 3 | 0 | 3 |
| GHITM | THAP2 | 0.692199 | 4 | 0 | 3 |
| GHITM | ZNF70 | 0.69022 | 3 | 0 | 3 |
| GHITM | TMEM19 | 0.689233 | 5 | 0 | 4 |
| GHITM | SYTL4 | 0.686374 | 3 | 0 | 3 |
| GHITM | CMTR2 | 0.685534 | 3 | 0 | 3 |
| GHITM | FYTTD1 | 0.683595 | 4 | 0 | 3 |
For details and further investigation, click here