| Full name: THAP domain containing 2 | Alias Symbol: DKFZP564I0422 | ||
| Type: protein-coding gene | Cytoband: 12q21.1 | ||
| Entrez ID: 83591 | HGNC ID: HGNC:20854 | Ensembl Gene: ENSG00000173451 | OMIM ID: 612531 |
| Drug and gene relationship at DGIdb | |||
Expression of THAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | THAP2 | 83591 | 230380_at | -0.4509 | 0.7193 | |
| GSE26886 | THAP2 | 83591 | 230380_at | -1.2176 | 0.0000 | |
| GSE45670 | THAP2 | 83591 | 230380_at | 0.0243 | 0.9515 | |
| GSE53622 | THAP2 | 83591 | 87986 | -0.4391 | 0.0114 | |
| GSE53624 | THAP2 | 83591 | 87986 | -0.2847 | 0.0715 | |
| GSE63941 | THAP2 | 83591 | 230380_at | -0.4236 | 0.5027 | |
| GSE77861 | THAP2 | 83591 | 230380_at | -0.4529 | 0.0408 | |
| GSE97050 | THAP2 | 83591 | A_24_P350437 | -0.1810 | 0.5659 | |
| SRP007169 | THAP2 | 83591 | RNAseq | 0.3615 | 0.5273 | |
| SRP064894 | THAP2 | 83591 | RNAseq | 0.0505 | 0.7993 | |
| SRP133303 | THAP2 | 83591 | RNAseq | 0.4093 | 0.0765 | |
| SRP159526 | THAP2 | 83591 | RNAseq | 0.1242 | 0.7275 | |
| SRP193095 | THAP2 | 83591 | RNAseq | -0.2454 | 0.1618 | |
| SRP219564 | THAP2 | 83591 | RNAseq | -0.4820 | 0.1009 | |
| TCGA | THAP2 | 83591 | RNAseq | -0.1525 | 0.1471 |
Upregulated datasets: 0; Downregulated datasets: 1.
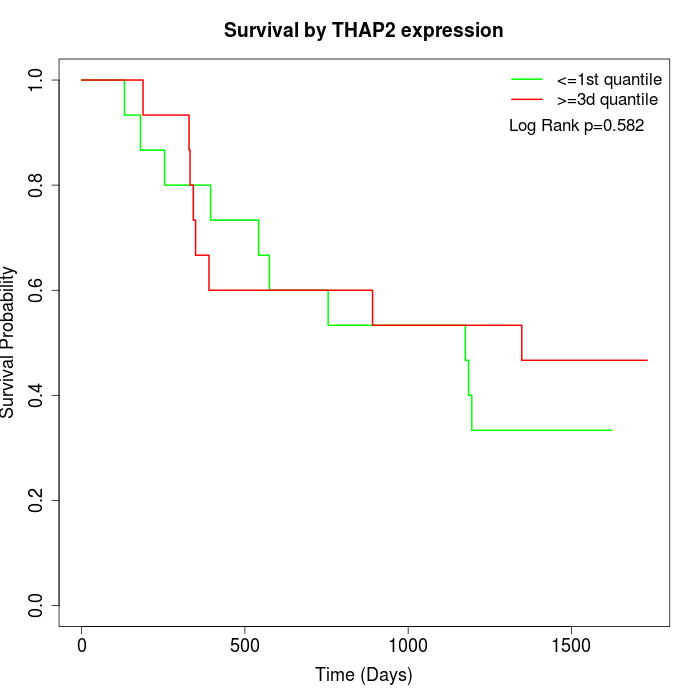
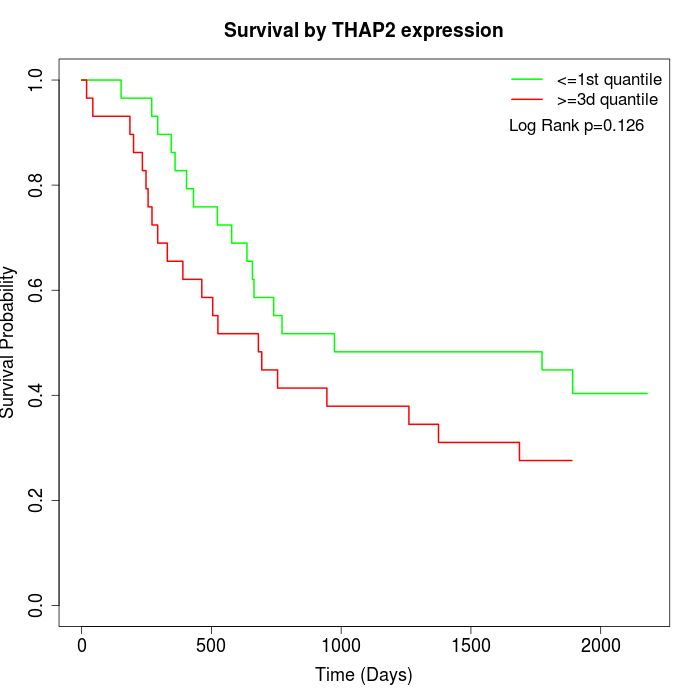
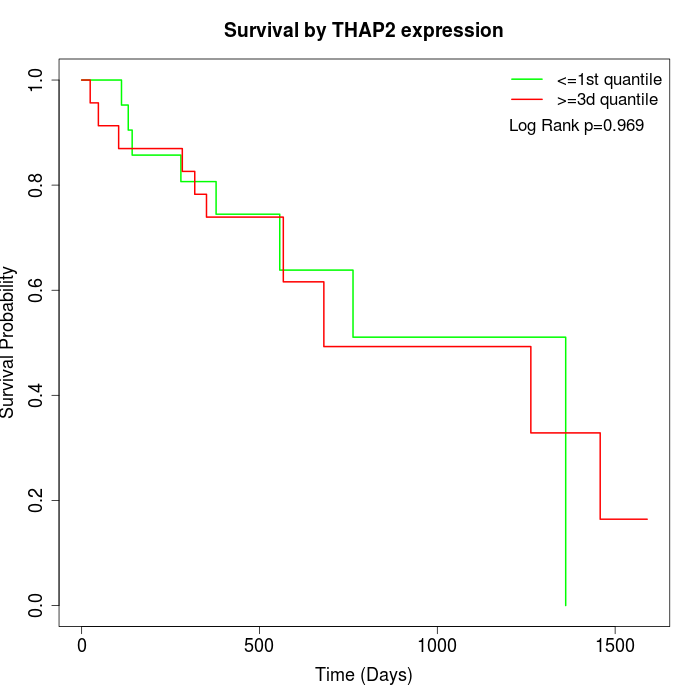
Survival by THAP2 expression:
Note: Click image to view full size file.
Copy number change of THAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | THAP2 | 83591 | 4 | 3 | 23 | |
| GSE20123 | THAP2 | 83591 | 4 | 3 | 23 | |
| GSE43470 | THAP2 | 83591 | 4 | 0 | 39 | |
| GSE46452 | THAP2 | 83591 | 8 | 1 | 50 | |
| GSE47630 | THAP2 | 83591 | 10 | 1 | 29 | |
| GSE54993 | THAP2 | 83591 | 0 | 5 | 65 | |
| GSE54994 | THAP2 | 83591 | 8 | 1 | 44 | |
| GSE60625 | THAP2 | 83591 | 0 | 0 | 11 | |
| GSE74703 | THAP2 | 83591 | 4 | 0 | 32 | |
| GSE74704 | THAP2 | 83591 | 3 | 2 | 15 | |
| TCGA | THAP2 | 83591 | 21 | 9 | 66 |
Total number of gains: 66; Total number of losses: 25; Total Number of normals: 397.
Somatic mutations of THAP2:
Generating mutation plots.
Highly correlated genes for THAP2:
Showing top 20/1055 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| THAP2 | CCDC25 | 0.830344 | 3 | 0 | 3 |
| THAP2 | ARL8B | 0.820202 | 3 | 0 | 3 |
| THAP2 | IMPACT | 0.804578 | 3 | 0 | 3 |
| THAP2 | ECHS1 | 0.80354 | 3 | 0 | 3 |
| THAP2 | ALDOA | 0.796556 | 3 | 0 | 3 |
| THAP2 | MGST2 | 0.79349 | 3 | 0 | 3 |
| THAP2 | GPD1L | 0.782022 | 3 | 0 | 3 |
| THAP2 | EIF4E | 0.781847 | 4 | 0 | 4 |
| THAP2 | MBNL3 | 0.78021 | 3 | 0 | 3 |
| THAP2 | TRIM23 | 0.780162 | 3 | 0 | 3 |
| THAP2 | TWF1 | 0.778347 | 4 | 0 | 4 |
| THAP2 | PITPNA | 0.774657 | 4 | 0 | 4 |
| THAP2 | KCTD10 | 0.768949 | 4 | 0 | 4 |
| THAP2 | ZNF717 | 0.768266 | 3 | 0 | 3 |
| THAP2 | HOXA4 | 0.766952 | 3 | 0 | 3 |
| THAP2 | ERI2 | 0.76537 | 3 | 0 | 3 |
| THAP2 | CARHSP1 | 0.762514 | 4 | 0 | 4 |
| THAP2 | TYW3 | 0.761406 | 3 | 0 | 3 |
| THAP2 | GOT1 | 0.761349 | 3 | 0 | 3 |
| THAP2 | EFCAB7 | 0.76098 | 3 | 0 | 3 |
For details and further investigation, click here