| Full name: glyoxalase domain containing 5 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp11.23 | ||
| Entrez ID: 392465 | HGNC ID: HGNC:33358 | Ensembl Gene: ENSG00000171433 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GLOD5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLOD5 | 392465 | 231052_at | 0.1602 | 0.4596 | |
| GSE26886 | GLOD5 | 392465 | 231052_at | 0.0500 | 0.7660 | |
| GSE45670 | GLOD5 | 392465 | 231052_at | 0.0330 | 0.8075 | |
| GSE53622 | GLOD5 | 392465 | 11250 | -0.2819 | 0.0016 | |
| GSE53624 | GLOD5 | 392465 | 11250 | -0.7857 | 0.0000 | |
| GSE63941 | GLOD5 | 392465 | 231052_at | 0.2759 | 0.1088 | |
| GSE77861 | GLOD5 | 392465 | 231052_at | -0.0710 | 0.4909 | |
| TCGA | GLOD5 | 392465 | RNAseq | -2.6682 | 0.0035 |
Upregulated datasets: 0; Downregulated datasets: 1.
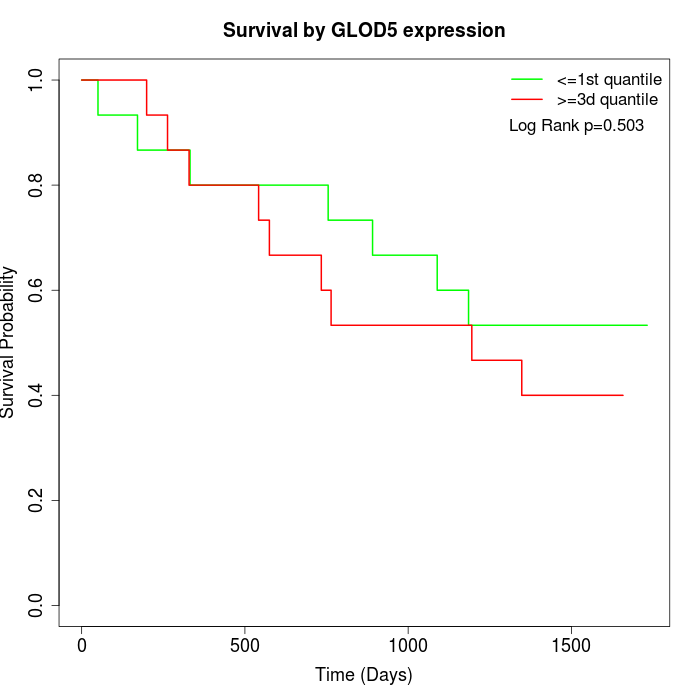
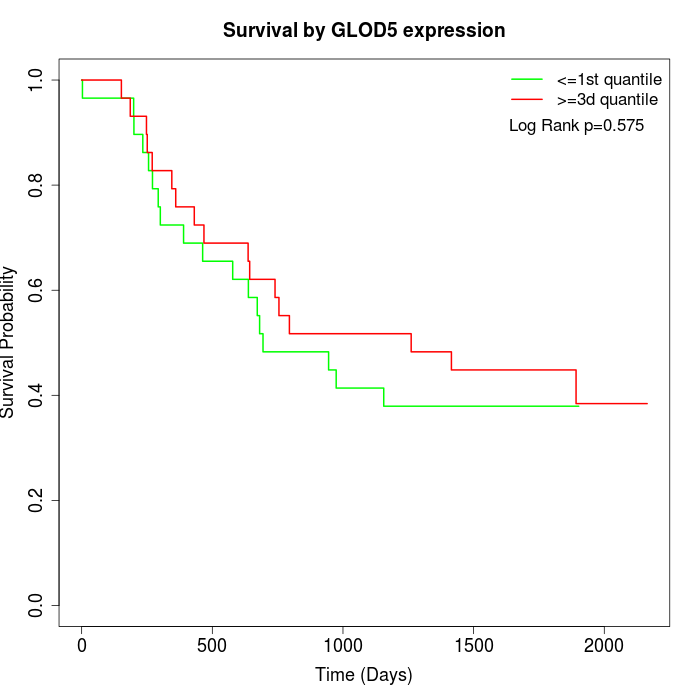
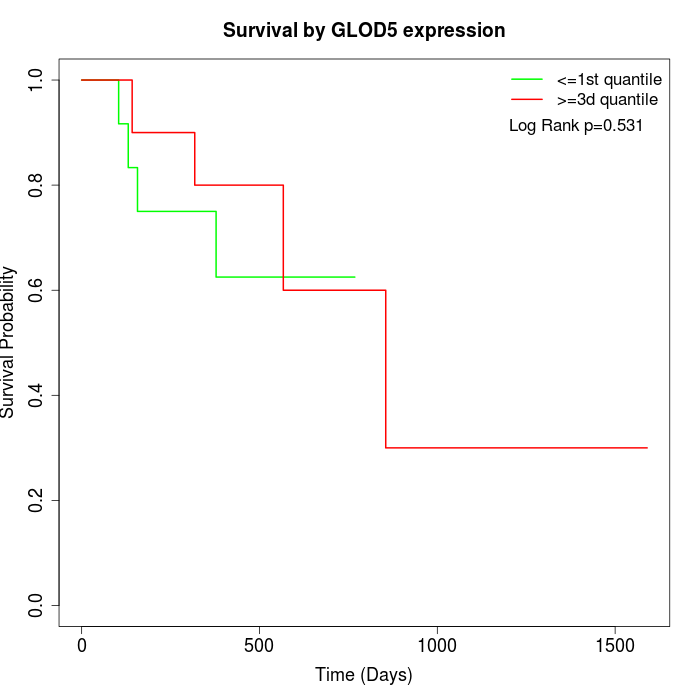
Survival by GLOD5 expression:
Note: Click image to view full size file.
Copy number change of GLOD5:
No record found for this gene.
Somatic mutations of GLOD5:
Generating mutation plots.
Highly correlated genes for GLOD5:
Showing top 20/50 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLOD5 | ONECUT2 | 0.63496 | 3 | 0 | 3 |
| GLOD5 | TTLL6 | 0.634258 | 3 | 0 | 3 |
| GLOD5 | CST8 | 0.616939 | 4 | 0 | 3 |
| GLOD5 | FOXH1 | 0.60158 | 3 | 0 | 3 |
| GLOD5 | KLF14 | 0.597533 | 3 | 0 | 3 |
| GLOD5 | GATA1 | 0.593507 | 4 | 0 | 3 |
| GLOD5 | CCDC182 | 0.585642 | 3 | 0 | 3 |
| GLOD5 | KCNIP2-AS1 | 0.580398 | 4 | 0 | 3 |
| GLOD5 | LINC01015 | 0.579216 | 4 | 0 | 3 |
| GLOD5 | PKD1L2 | 0.579107 | 4 | 0 | 3 |
| GLOD5 | TMPRSS6 | 0.578908 | 4 | 0 | 3 |
| GLOD5 | PRB1 | 0.577761 | 4 | 0 | 3 |
| GLOD5 | MOGAT2 | 0.57566 | 4 | 0 | 3 |
| GLOD5 | VGLL1 | 0.574486 | 5 | 0 | 3 |
| GLOD5 | HPX | 0.574481 | 4 | 0 | 3 |
| GLOD5 | PTGES2 | 0.568679 | 3 | 0 | 3 |
| GLOD5 | CD180 | 0.566013 | 3 | 0 | 3 |
| GLOD5 | TEX38 | 0.563311 | 4 | 0 | 3 |
| GLOD5 | LYL1 | 0.561141 | 3 | 0 | 3 |
| GLOD5 | SRCIN1 | 0.555674 | 4 | 0 | 3 |
For details and further investigation, click here