| Full name: polycystin 1 like 2 (gene/pseudogene) | Alias Symbol: KIAA1879 | ||
| Type: protein-coding gene | Cytoband: 16q23.2 | ||
| Entrez ID: 114780 | HGNC ID: HGNC:21715 | Ensembl Gene: ENSG00000166473 | OMIM ID: 607894 |
| Drug and gene relationship at DGIdb | |||
Expression of PKD1L2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PKD1L2 | 114780 | 1559261_a_at | -0.0866 | 0.7346 | |
| GSE26886 | PKD1L2 | 114780 | 1559261_a_at | 0.0991 | 0.4024 | |
| GSE45670 | PKD1L2 | 114780 | 1559261_a_at | 0.0689 | 0.6121 | |
| GSE53622 | PKD1L2 | 114780 | 85675 | -1.1419 | 0.0000 | |
| GSE53624 | PKD1L2 | 114780 | 85675 | -2.0494 | 0.0000 | |
| GSE63941 | PKD1L2 | 114780 | 1559261_a_at | 0.3686 | 0.1999 | |
| GSE77861 | PKD1L2 | 114780 | 1559261_a_at | -0.0880 | 0.3433 | |
| GSE97050 | PKD1L2 | 114780 | A_24_P246518 | -0.8252 | 0.1187 | |
| SRP133303 | PKD1L2 | 114780 | RNAseq | -1.8986 | 0.0000 | |
| SRP159526 | PKD1L2 | 114780 | RNAseq | -0.5979 | 0.5747 | |
| TCGA | PKD1L2 | 114780 | RNAseq | -0.3131 | 0.5601 |
Upregulated datasets: 0; Downregulated datasets: 3.
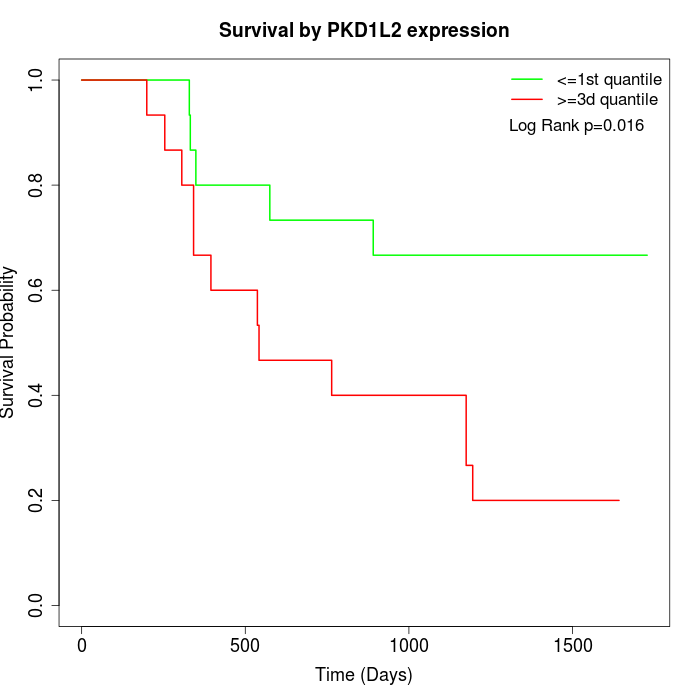
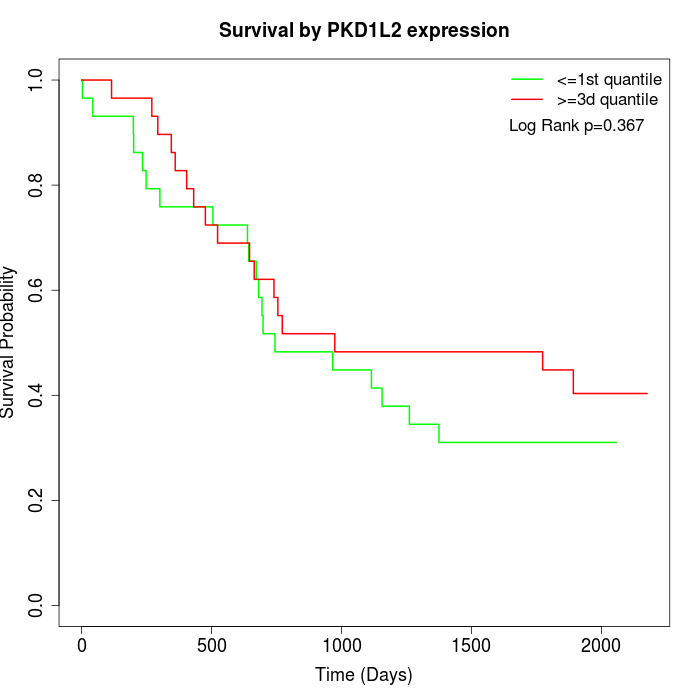
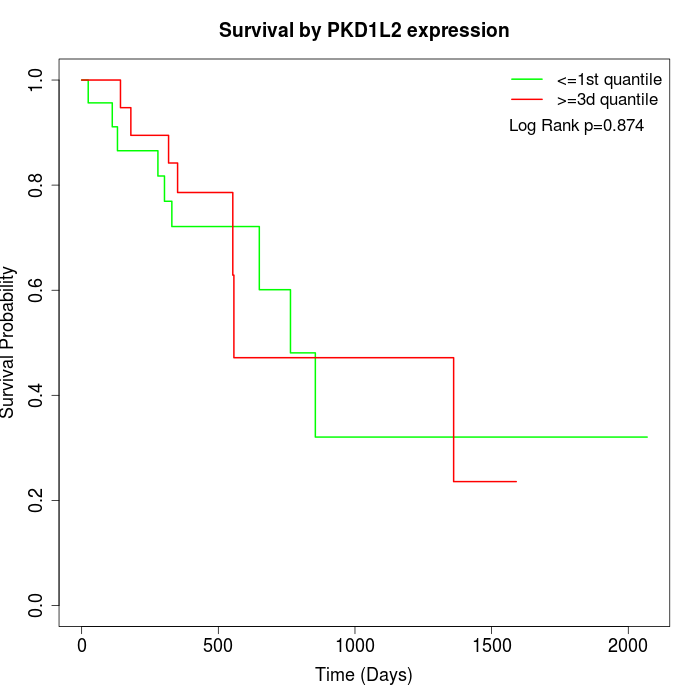
Survival by PKD1L2 expression:
Note: Click image to view full size file.
Copy number change of PKD1L2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PKD1L2 | 114780 | 5 | 3 | 22 | |
| GSE20123 | PKD1L2 | 114780 | 5 | 3 | 22 | |
| GSE43470 | PKD1L2 | 114780 | 2 | 8 | 33 | |
| GSE46452 | PKD1L2 | 114780 | 37 | 2 | 20 | |
| GSE47630 | PKD1L2 | 114780 | 11 | 8 | 21 | |
| GSE54993 | PKD1L2 | 114780 | 2 | 4 | 64 | |
| GSE54994 | PKD1L2 | 114780 | 7 | 10 | 36 | |
| GSE60625 | PKD1L2 | 114780 | 4 | 0 | 7 | |
| GSE74703 | PKD1L2 | 114780 | 2 | 6 | 28 | |
| GSE74704 | PKD1L2 | 114780 | 3 | 2 | 15 | |
| TCGA | PKD1L2 | 114780 | 24 | 16 | 56 |
Total number of gains: 102; Total number of losses: 62; Total Number of normals: 324.
Somatic mutations of PKD1L2:
Generating mutation plots.
Highly correlated genes for PKD1L2:
Showing top 20/172 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PKD1L2 | BOK | 0.750335 | 3 | 0 | 3 |
| PKD1L2 | GJC1 | 0.696696 | 3 | 0 | 3 |
| PKD1L2 | VMAC | 0.695938 | 3 | 0 | 3 |
| PKD1L2 | LACTBL1 | 0.693482 | 3 | 0 | 3 |
| PKD1L2 | KRTAP5-10 | 0.685128 | 3 | 0 | 3 |
| PKD1L2 | ARHGEF10L | 0.684925 | 3 | 0 | 3 |
| PKD1L2 | PEBP1 | 0.68249 | 3 | 0 | 3 |
| PKD1L2 | HAR1A | 0.682314 | 3 | 0 | 3 |
| PKD1L2 | SPATA31E1 | 0.681798 | 3 | 0 | 3 |
| PKD1L2 | KRT78 | 0.670399 | 3 | 0 | 3 |
| PKD1L2 | CDX1 | 0.661319 | 5 | 0 | 4 |
| PKD1L2 | DSTN | 0.655297 | 3 | 0 | 3 |
| PKD1L2 | PTCH2 | 0.649015 | 3 | 0 | 3 |
| PKD1L2 | CLNK | 0.648787 | 3 | 0 | 3 |
| PKD1L2 | EFNA3 | 0.648229 | 3 | 0 | 3 |
| PKD1L2 | PHACTR4 | 0.6456 | 3 | 0 | 3 |
| PKD1L2 | SORBS1 | 0.643639 | 3 | 0 | 3 |
| PKD1L2 | RPL32 | 0.642759 | 3 | 0 | 3 |
| PKD1L2 | HPGD | 0.642559 | 3 | 0 | 3 |
| PKD1L2 | KDM5C | 0.64189 | 3 | 0 | 3 |
For details and further investigation, click here