| Full name: prostaglandin E synthase 2 | Alias Symbol: FLJ14038 | ||
| Type: protein-coding gene | Cytoband: 9q34.11 | ||
| Entrez ID: 80142 | HGNC ID: HGNC:17822 | Ensembl Gene: ENSG00000148334 | OMIM ID: 608152 |
| Drug and gene relationship at DGIdb | |||
Expression of PTGES2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PTGES2 | 80142 | 218083_at | 0.1870 | 0.4385 | |
| GSE20347 | PTGES2 | 80142 | 218083_at | 0.0422 | 0.6968 | |
| GSE23400 | PTGES2 | 80142 | 218083_at | 0.1400 | 0.0234 | |
| GSE26886 | PTGES2 | 80142 | 218083_at | -0.3258 | 0.1432 | |
| GSE29001 | PTGES2 | 80142 | 218083_at | 0.2024 | 0.4213 | |
| GSE38129 | PTGES2 | 80142 | 218083_at | 0.1259 | 0.0912 | |
| GSE45670 | PTGES2 | 80142 | 218083_at | 0.1334 | 0.1760 | |
| GSE53622 | PTGES2 | 80142 | 111758 | 0.0641 | 0.4329 | |
| GSE53624 | PTGES2 | 80142 | 111758 | 0.3391 | 0.0000 | |
| GSE63941 | PTGES2 | 80142 | 218083_at | 1.1940 | 0.0010 | |
| GSE77861 | PTGES2 | 80142 | 218083_at | 0.2036 | 0.1360 | |
| GSE97050 | PTGES2 | 80142 | A_24_P106953 | 0.1546 | 0.7123 | |
| SRP007169 | PTGES2 | 80142 | RNAseq | 0.0499 | 0.9119 | |
| SRP008496 | PTGES2 | 80142 | RNAseq | -0.0457 | 0.8483 | |
| SRP064894 | PTGES2 | 80142 | RNAseq | 0.5637 | 0.0597 | |
| SRP133303 | PTGES2 | 80142 | RNAseq | 0.3082 | 0.0738 | |
| SRP159526 | PTGES2 | 80142 | RNAseq | 0.3723 | 0.0367 | |
| SRP193095 | PTGES2 | 80142 | RNAseq | 0.1896 | 0.1390 | |
| SRP219564 | PTGES2 | 80142 | RNAseq | -0.0628 | 0.8410 | |
| TCGA | PTGES2 | 80142 | RNAseq | 0.0742 | 0.1747 |
Upregulated datasets: 1; Downregulated datasets: 0.
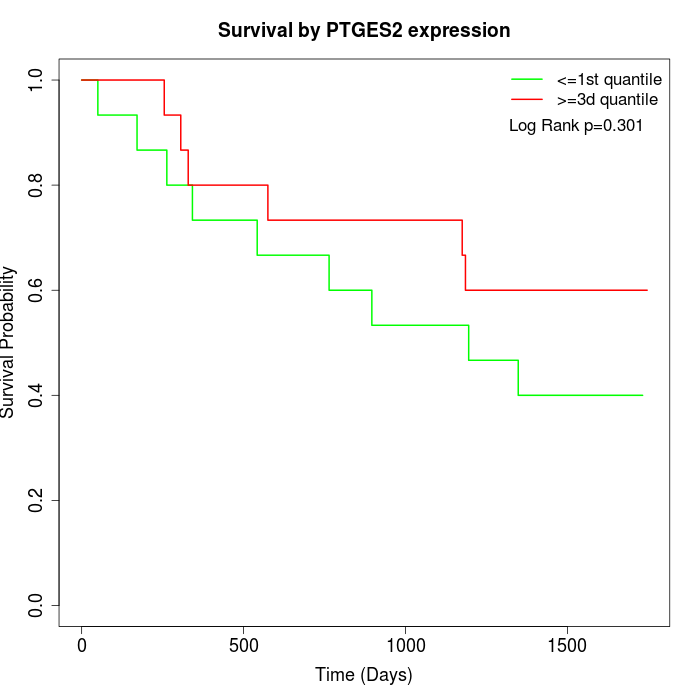
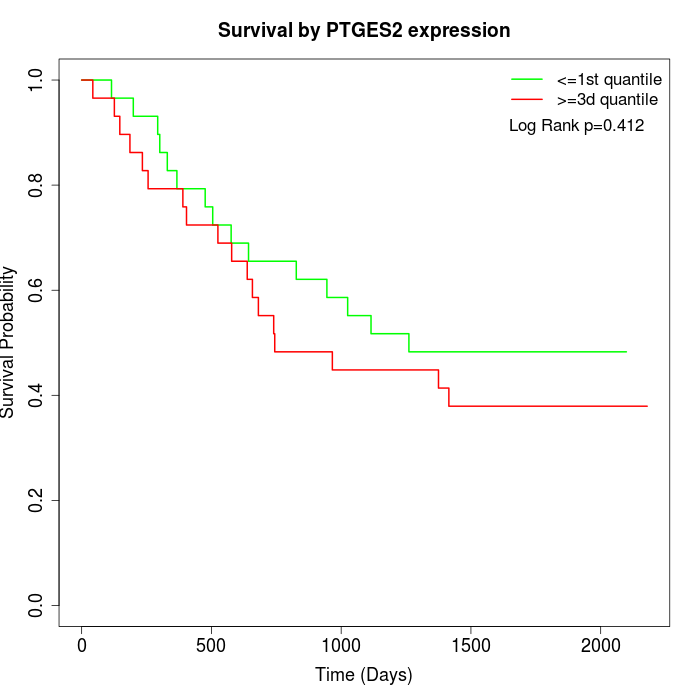
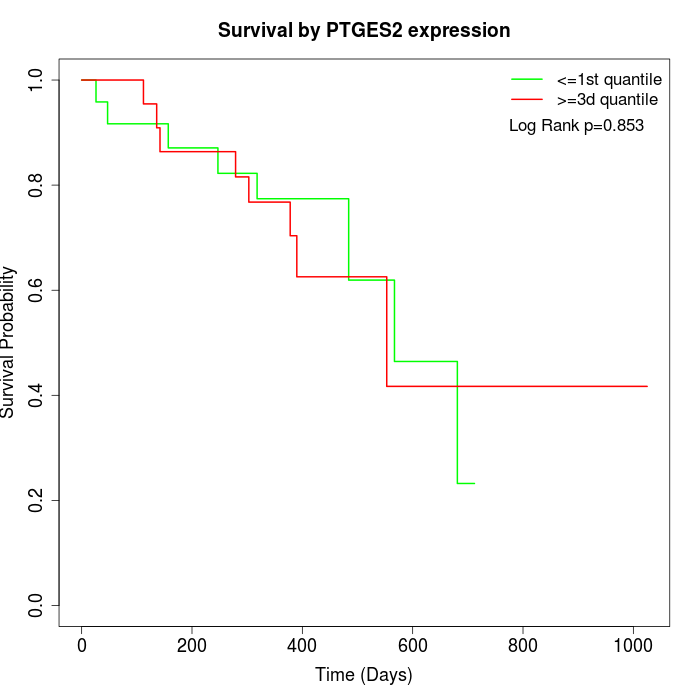
Survival by PTGES2 expression:
Note: Click image to view full size file.
Copy number change of PTGES2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PTGES2 | 80142 | 6 | 9 | 15 | |
| GSE20123 | PTGES2 | 80142 | 6 | 9 | 15 | |
| GSE43470 | PTGES2 | 80142 | 5 | 7 | 31 | |
| GSE46452 | PTGES2 | 80142 | 6 | 13 | 40 | |
| GSE47630 | PTGES2 | 80142 | 3 | 16 | 21 | |
| GSE54993 | PTGES2 | 80142 | 3 | 3 | 64 | |
| GSE54994 | PTGES2 | 80142 | 11 | 8 | 34 | |
| GSE60625 | PTGES2 | 80142 | 0 | 0 | 11 | |
| GSE74703 | PTGES2 | 80142 | 5 | 5 | 26 | |
| GSE74704 | PTGES2 | 80142 | 3 | 7 | 10 | |
| TCGA | PTGES2 | 80142 | 27 | 25 | 44 |
Total number of gains: 75; Total number of losses: 102; Total Number of normals: 311.
Somatic mutations of PTGES2:
Generating mutation plots.
Highly correlated genes for PTGES2:
Showing top 20/633 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PTGES2 | MPV17L | 0.77918 | 3 | 0 | 3 |
| PTGES2 | WBP2 | 0.775442 | 3 | 0 | 3 |
| PTGES2 | DLL4 | 0.764961 | 3 | 0 | 3 |
| PTGES2 | C12orf45 | 0.748448 | 3 | 0 | 3 |
| PTGES2 | PRELID1 | 0.746309 | 4 | 0 | 4 |
| PTGES2 | PPP5C | 0.738141 | 6 | 0 | 5 |
| PTGES2 | GIGYF1 | 0.738051 | 3 | 0 | 3 |
| PTGES2 | GHDC | 0.728813 | 3 | 0 | 3 |
| PTGES2 | FBXO16 | 0.727666 | 3 | 0 | 3 |
| PTGES2 | CLK2 | 0.723574 | 3 | 0 | 3 |
| PTGES2 | KIAA1257 | 0.720864 | 3 | 0 | 3 |
| PTGES2 | NIT1 | 0.715991 | 3 | 0 | 3 |
| PTGES2 | GNB1 | 0.715405 | 3 | 0 | 3 |
| PTGES2 | AAMP | 0.710924 | 4 | 0 | 4 |
| PTGES2 | RFTN2 | 0.709374 | 4 | 0 | 4 |
| PTGES2 | AP1M2 | 0.703903 | 5 | 0 | 4 |
| PTGES2 | HOMER2 | 0.70134 | 3 | 0 | 3 |
| PTGES2 | SLC35A4 | 0.699388 | 3 | 0 | 3 |
| PTGES2 | DENND4B | 0.69859 | 3 | 0 | 3 |
| PTGES2 | MSH2 | 0.696108 | 3 | 0 | 3 |
For details and further investigation, click here