| Full name: glycosylphosphatidylinositol anchored molecule like | Alias Symbol: LY6DL | ||
| Type: protein-coding gene | Cytoband: 8q24.3 | ||
| Entrez ID: 2765 | HGNC ID: HGNC:4375 | Ensembl Gene: ENSG00000104499 | OMIM ID: 602370 |
| Drug and gene relationship at DGIdb | |||
Expression of GML:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GML | 2765 | 208000_at | -0.0194 | 0.9602 | |
| GSE20347 | GML | 2765 | 208000_at | 0.0427 | 0.6004 | |
| GSE23400 | GML | 2765 | 208000_at | -0.1116 | 0.0360 | |
| GSE26886 | GML | 2765 | 208000_at | -0.0885 | 0.5690 | |
| GSE29001 | GML | 2765 | 208000_at | -0.2082 | 0.3049 | |
| GSE38129 | GML | 2765 | 208000_at | -0.0795 | 0.4504 | |
| GSE45670 | GML | 2765 | 208000_at | 0.0785 | 0.3840 | |
| GSE53622 | GML | 2765 | 30954 | -0.0011 | 0.9891 | |
| GSE53624 | GML | 2765 | 30954 | -0.1615 | 0.0208 | |
| GSE63941 | GML | 2765 | 208000_at | 0.2720 | 0.0378 | |
| GSE77861 | GML | 2765 | 208000_at | -0.1768 | 0.3489 |
Upregulated datasets: 0; Downregulated datasets: 0.
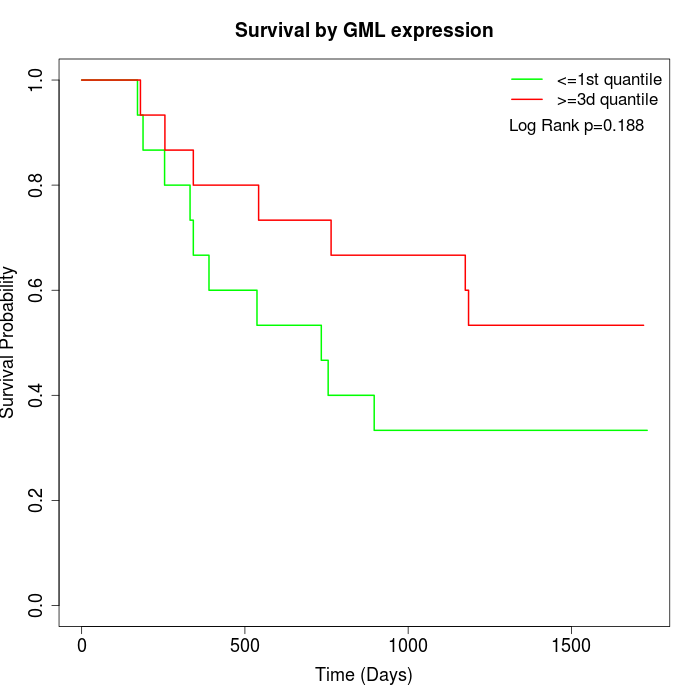
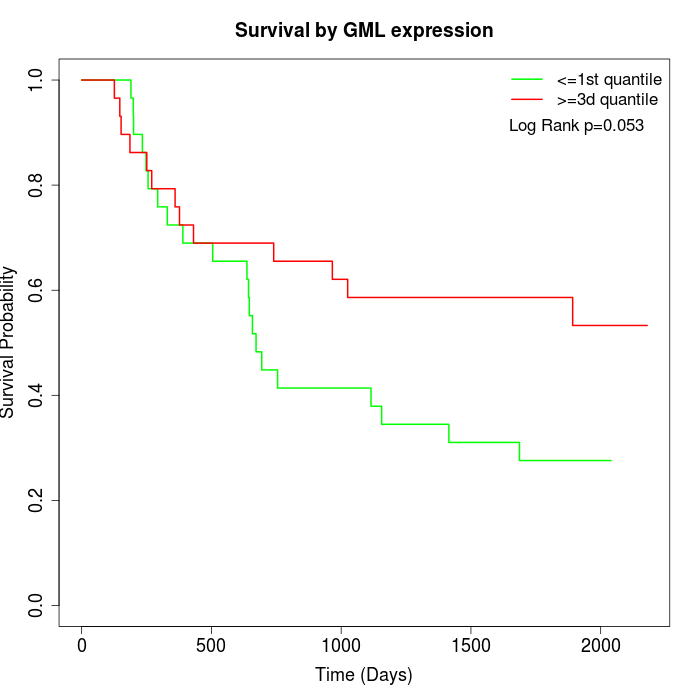
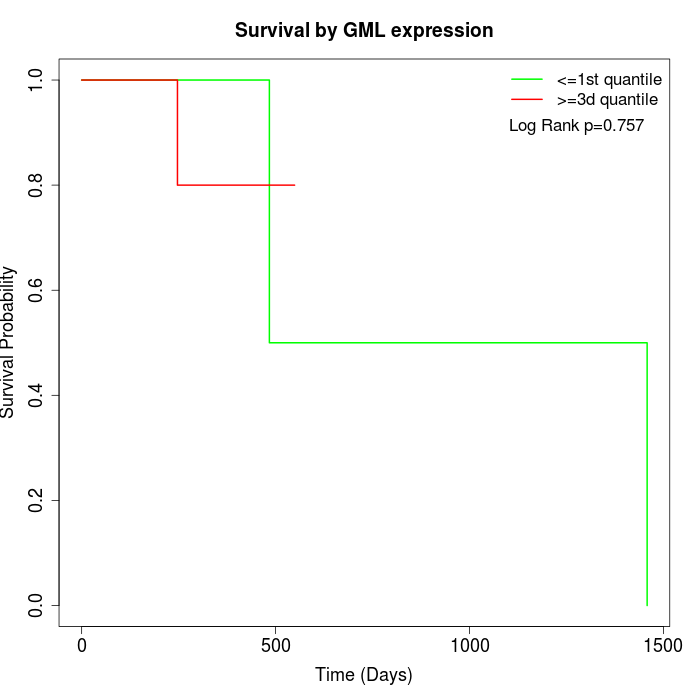
Survival by GML expression:
Note: Click image to view full size file.
Copy number change of GML:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GML | 2765 | 16 | 1 | 13 | |
| GSE20123 | GML | 2765 | 17 | 1 | 12 | |
| GSE43470 | GML | 2765 | 17 | 2 | 24 | |
| GSE46452 | GML | 2765 | 27 | 0 | 32 | |
| GSE47630 | GML | 2765 | 23 | 1 | 16 | |
| GSE54993 | GML | 2765 | 0 | 25 | 45 | |
| GSE54994 | GML | 2765 | 39 | 1 | 13 | |
| GSE60625 | GML | 2765 | 3 | 4 | 4 | |
| GSE74703 | GML | 2765 | 15 | 1 | 20 | |
| GSE74704 | GML | 2765 | 10 | 0 | 10 | |
| TCGA | GML | 2765 | 64 | 2 | 30 |
Total number of gains: 231; Total number of losses: 38; Total Number of normals: 219.
Somatic mutations of GML:
Generating mutation plots.
Highly correlated genes for GML:
Showing top 20/1024 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GML | CBL | 0.778255 | 4 | 0 | 4 |
| GML | CIITA | 0.759344 | 5 | 0 | 5 |
| GML | IL1RAPL2 | 0.758219 | 6 | 0 | 6 |
| GML | KHDRBS2 | 0.756739 | 5 | 0 | 5 |
| GML | ECE2 | 0.739544 | 4 | 0 | 4 |
| GML | ACSBG2 | 0.731249 | 6 | 0 | 6 |
| GML | CC2D2B | 0.728365 | 3 | 0 | 3 |
| GML | LUZP4 | 0.727677 | 6 | 0 | 6 |
| GML | FCGR2C | 0.718672 | 3 | 0 | 3 |
| GML | TRPM3 | 0.718413 | 5 | 0 | 5 |
| GML | NEUROD2 | 0.71775 | 5 | 0 | 5 |
| GML | PKD2L2 | 0.716106 | 3 | 0 | 3 |
| GML | FABP1 | 0.715896 | 3 | 0 | 3 |
| GML | LBP | 0.710617 | 5 | 0 | 5 |
| GML | DPF3 | 0.704516 | 5 | 0 | 5 |
| GML | IL21 | 0.703485 | 5 | 0 | 5 |
| GML | RSPH6A | 0.703358 | 4 | 0 | 4 |
| GML | CYP1A2 | 0.702407 | 5 | 0 | 5 |
| GML | OPRM1 | 0.701208 | 6 | 0 | 6 |
| GML | CCDC7 | 0.700912 | 4 | 0 | 4 |
For details and further investigation, click here