| Full name: G protein-coupled receptor 143 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp22.2 | ||
| Entrez ID: 4935 | HGNC ID: HGNC:20145 | Ensembl Gene: ENSG00000101850 | OMIM ID: 300808 |
| Drug and gene relationship at DGIdb | |||
Expression of GPR143:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPR143 | 4935 | 206696_at | 0.5797 | 0.6758 | |
| GSE20347 | GPR143 | 4935 | 206696_at | -0.2160 | 0.0302 | |
| GSE23400 | GPR143 | 4935 | 206696_at | -0.0879 | 0.1480 | |
| GSE26886 | GPR143 | 4935 | 206696_at | 0.1240 | 0.7196 | |
| GSE29001 | GPR143 | 4935 | 206696_at | -0.1928 | 0.4145 | |
| GSE38129 | GPR143 | 4935 | 206696_at | -0.0269 | 0.8458 | |
| GSE45670 | GPR143 | 4935 | 206696_at | 0.1519 | 0.5446 | |
| GSE53622 | GPR143 | 4935 | 116441 | -0.4737 | 0.0376 | |
| GSE53624 | GPR143 | 4935 | 116441 | -1.0082 | 0.0000 | |
| GSE63941 | GPR143 | 4935 | 206696_at | 0.6406 | 0.3973 | |
| GSE77861 | GPR143 | 4935 | 206696_at | -0.0782 | 0.6010 | |
| GSE97050 | GPR143 | 4935 | A_23_P45560 | 0.3269 | 0.3200 | |
| SRP064894 | GPR143 | 4935 | RNAseq | -1.2776 | 0.0001 | |
| SRP133303 | GPR143 | 4935 | RNAseq | -0.6501 | 0.0099 | |
| SRP159526 | GPR143 | 4935 | RNAseq | 0.3995 | 0.6178 | |
| SRP219564 | GPR143 | 4935 | RNAseq | -0.6949 | 0.3630 | |
| TCGA | GPR143 | 4935 | RNAseq | 0.4755 | 0.2196 |
Upregulated datasets: 0; Downregulated datasets: 2.
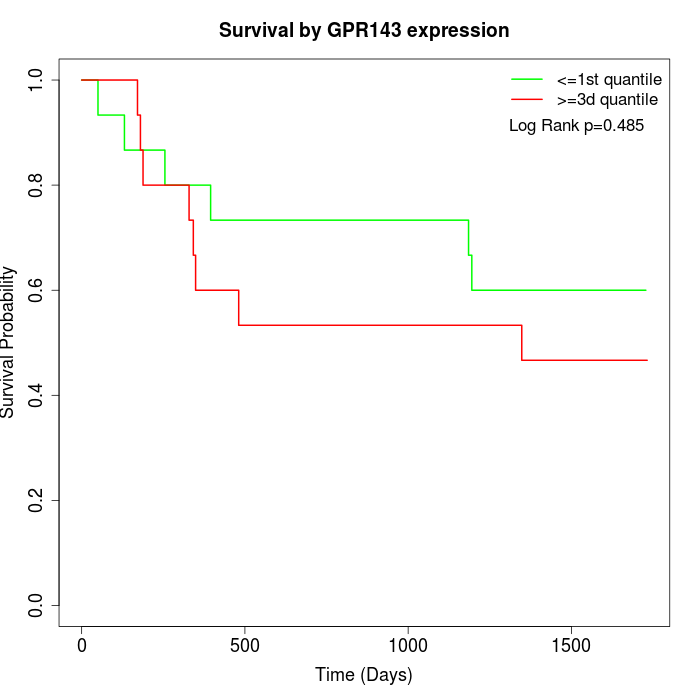
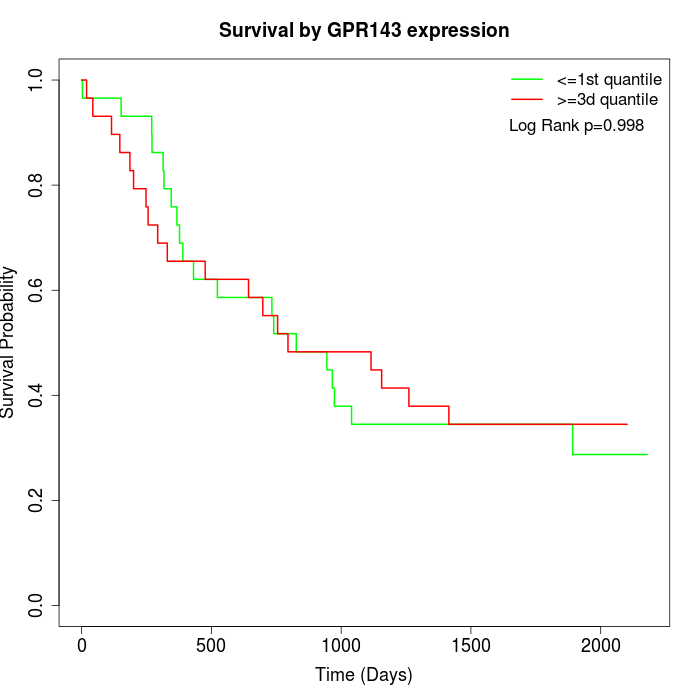
Survival by GPR143 expression:
Note: Click image to view full size file.
Copy number change of GPR143:
No record found for this gene.
Somatic mutations of GPR143:
Generating mutation plots.
Highly correlated genes for GPR143:
Showing top 20/153 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPR143 | C6orf136 | 0.761006 | 3 | 0 | 3 |
| GPR143 | RNASEH2B | 0.694101 | 3 | 0 | 3 |
| GPR143 | ZSCAN16 | 0.688961 | 3 | 0 | 3 |
| GPR143 | ABHD16A | 0.688021 | 3 | 0 | 3 |
| GPR143 | TMEM69 | 0.686563 | 3 | 0 | 3 |
| GPR143 | SLAIN1 | 0.684559 | 3 | 0 | 3 |
| GPR143 | ZNF765 | 0.675959 | 3 | 0 | 3 |
| GPR143 | STAMBPL1 | 0.675608 | 3 | 0 | 3 |
| GPR143 | ZNF814 | 0.665014 | 3 | 0 | 3 |
| GPR143 | UBP1 | 0.663343 | 3 | 0 | 3 |
| GPR143 | YIPF3 | 0.660964 | 4 | 0 | 3 |
| GPR143 | PTPRA | 0.6552 | 3 | 0 | 3 |
| GPR143 | PIK3R3 | 0.647409 | 4 | 0 | 4 |
| GPR143 | ABHD12 | 0.641816 | 3 | 0 | 3 |
| GPR143 | LMLN | 0.637819 | 3 | 0 | 3 |
| GPR143 | ZNF138 | 0.635507 | 3 | 0 | 3 |
| GPR143 | HAVCR2 | 0.635123 | 3 | 0 | 3 |
| GPR143 | SLC35A5 | 0.632443 | 3 | 0 | 3 |
| GPR143 | PPP1R11 | 0.631172 | 3 | 0 | 3 |
| GPR143 | C12orf73 | 0.630838 | 4 | 0 | 3 |
For details and further investigation, click here