| Full name: G protein-coupled receptor 62 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3p21.2 | ||
| Entrez ID: 118442 | HGNC ID: HGNC:13301 | Ensembl Gene: ENSG00000180929 | OMIM ID: 606917 |
| Drug and gene relationship at DGIdb | |||
Expression of GPR62:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPR62 | 118442 | 1554559_at | -0.1406 | 0.6356 | |
| GSE26886 | GPR62 | 118442 | 1554559_at | 0.1788 | 0.2454 | |
| GSE45670 | GPR62 | 118442 | 1554559_at | 0.0952 | 0.2275 | |
| GSE53622 | GPR62 | 118442 | 53920 | -0.6337 | 0.0000 | |
| GSE53624 | GPR62 | 118442 | 53920 | -1.1769 | 0.0000 | |
| GSE63941 | GPR62 | 118442 | 1554559_at | 0.1823 | 0.1814 | |
| GSE77861 | GPR62 | 118442 | 1554559_at | -0.0831 | 0.4871 | |
| GSE97050 | GPR62 | 118442 | A_23_P434289 | -0.0222 | 0.9469 | |
| TCGA | GPR62 | 118442 | RNAseq | -0.5597 | 0.1111 |
Upregulated datasets: 0; Downregulated datasets: 1.
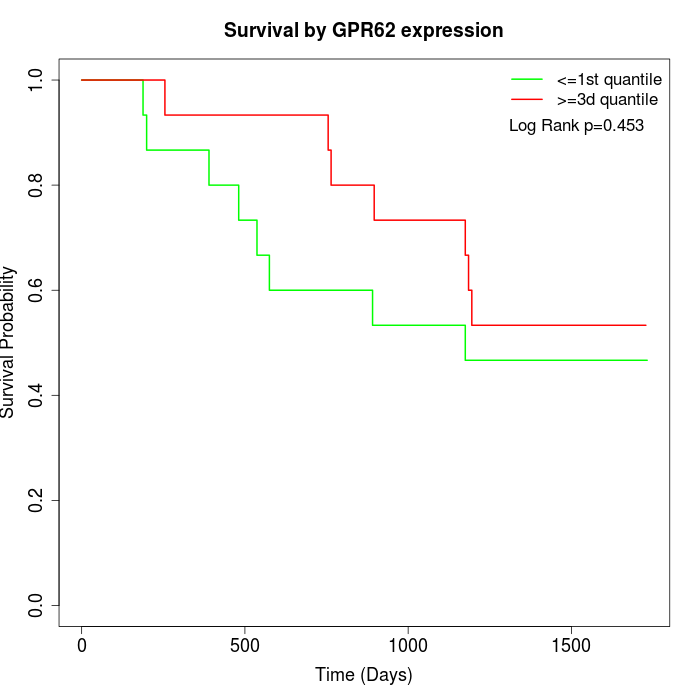
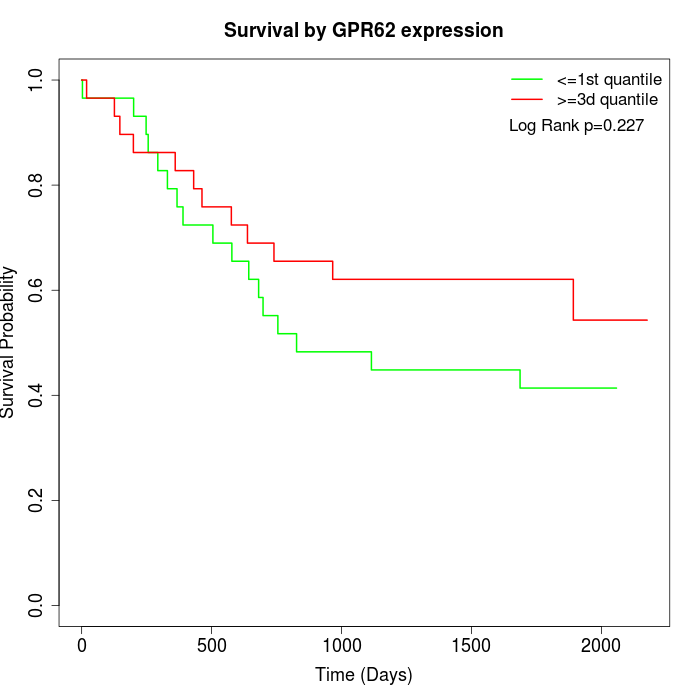
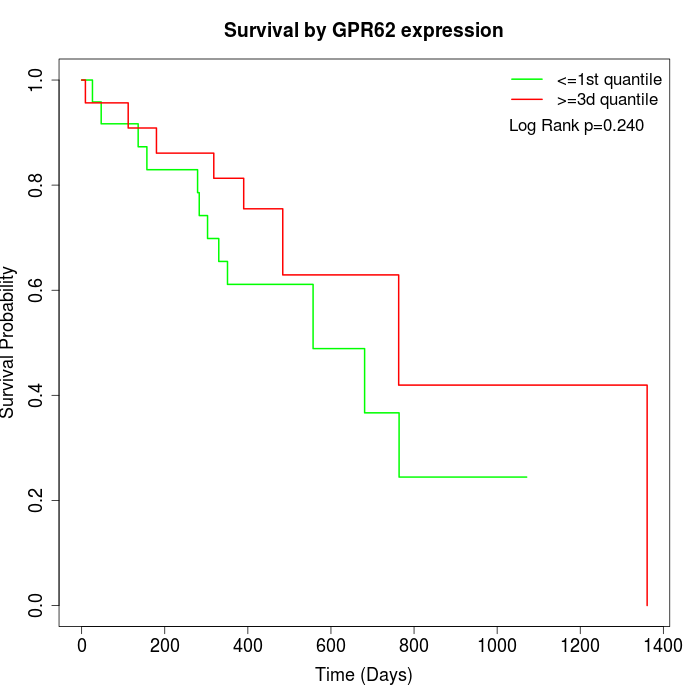
Survival by GPR62 expression:
Note: Click image to view full size file.
Copy number change of GPR62:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPR62 | 118442 | 0 | 18 | 12 | |
| GSE20123 | GPR62 | 118442 | 0 | 19 | 11 | |
| GSE43470 | GPR62 | 118442 | 0 | 18 | 25 | |
| GSE46452 | GPR62 | 118442 | 2 | 17 | 40 | |
| GSE47630 | GPR62 | 118442 | 1 | 24 | 15 | |
| GSE54993 | GPR62 | 118442 | 6 | 2 | 62 | |
| GSE54994 | GPR62 | 118442 | 0 | 34 | 19 | |
| GSE60625 | GPR62 | 118442 | 5 | 0 | 6 | |
| GSE74703 | GPR62 | 118442 | 0 | 14 | 22 | |
| GSE74704 | GPR62 | 118442 | 0 | 12 | 8 | |
| TCGA | GPR62 | 118442 | 0 | 78 | 18 |
Total number of gains: 14; Total number of losses: 236; Total Number of normals: 238.
Somatic mutations of GPR62:
Generating mutation plots.
Highly correlated genes for GPR62:
Showing top 20/272 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPR62 | KRTAP5-4 | 0.903659 | 3 | 0 | 3 |
| GPR62 | RNF222 | 0.879642 | 3 | 0 | 3 |
| GPR62 | CLEC18C | 0.8487 | 3 | 0 | 3 |
| GPR62 | HAMP | 0.835727 | 3 | 0 | 3 |
| GPR62 | KRTAP5-5 | 0.831679 | 3 | 0 | 3 |
| GPR62 | NOTO | 0.816285 | 3 | 0 | 3 |
| GPR62 | KRTAP5-11 | 0.813936 | 3 | 0 | 3 |
| GPR62 | BARHL2 | 0.803061 | 3 | 0 | 3 |
| GPR62 | KCNA7 | 0.796612 | 4 | 0 | 4 |
| GPR62 | KRTAP10-8 | 0.783986 | 3 | 0 | 3 |
| GPR62 | MYO1A | 0.771181 | 3 | 0 | 3 |
| GPR62 | ACBD4 | 0.767918 | 3 | 0 | 3 |
| GPR62 | CLEC12B | 0.765043 | 4 | 0 | 3 |
| GPR62 | ATP2B3 | 0.755573 | 5 | 0 | 4 |
| GPR62 | FAM163B | 0.755164 | 3 | 0 | 3 |
| GPR62 | MAFG | 0.754034 | 3 | 0 | 3 |
| GPR62 | IL37 | 0.742194 | 4 | 0 | 4 |
| GPR62 | ADAMTSL4 | 0.737724 | 3 | 0 | 3 |
| GPR62 | ADRA1A | 0.733541 | 3 | 0 | 3 |
| GPR62 | CD164L2 | 0.731972 | 3 | 0 | 3 |
For details and further investigation, click here