| Full name: hepcidin antimicrobial peptide | Alias Symbol: LEAP-1|HEPC|HFE2B|LEAP1 | ||
| Type: protein-coding gene | Cytoband: 19q13.12 | ||
| Entrez ID: 57817 | HGNC ID: HGNC:15598 | Ensembl Gene: ENSG00000105697 | OMIM ID: 606464 |
| Drug and gene relationship at DGIdb | |||
Expression of HAMP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HAMP | 57817 | 220491_at | 0.1954 | 0.3975 | |
| GSE20347 | HAMP | 57817 | 220491_at | 0.0617 | 0.4798 | |
| GSE23400 | HAMP | 57817 | 220491_at | -0.0696 | 0.0120 | |
| GSE26886 | HAMP | 57817 | 220491_at | -0.1822 | 0.3177 | |
| GSE29001 | HAMP | 57817 | 220491_at | 0.0594 | 0.6812 | |
| GSE38129 | HAMP | 57817 | 220491_at | 0.1582 | 0.1731 | |
| GSE45670 | HAMP | 57817 | 220491_at | 0.0317 | 0.8342 | |
| GSE53622 | HAMP | 57817 | 40953 | -1.2733 | 0.0000 | |
| GSE53624 | HAMP | 57817 | 40953 | -1.5538 | 0.0000 | |
| GSE63941 | HAMP | 57817 | 220491_at | 0.0429 | 0.8513 | |
| GSE77861 | HAMP | 57817 | 220491_at | -0.2055 | 0.1019 | |
| GSE97050 | HAMP | 57817 | A_33_P3336720 | -0.0709 | 0.8401 | |
| TCGA | HAMP | 57817 | RNAseq | 1.5411 | 0.0150 |
Upregulated datasets: 1; Downregulated datasets: 2.
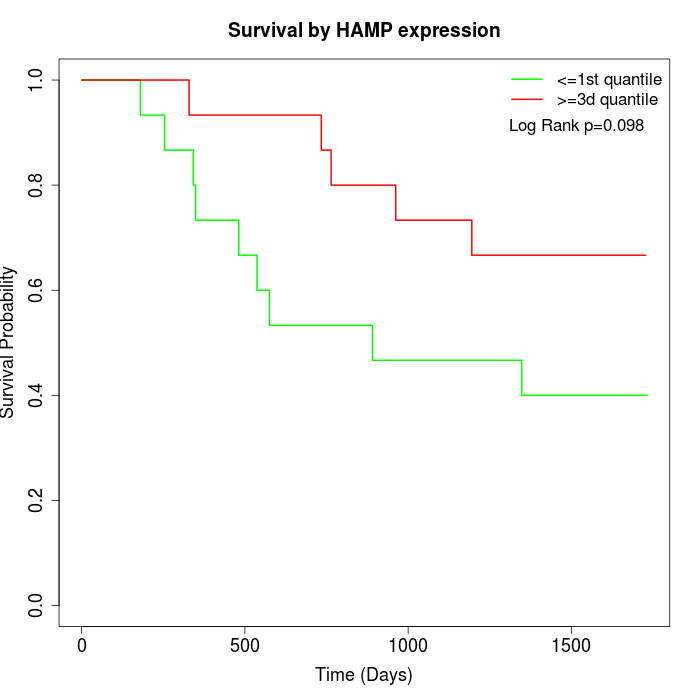
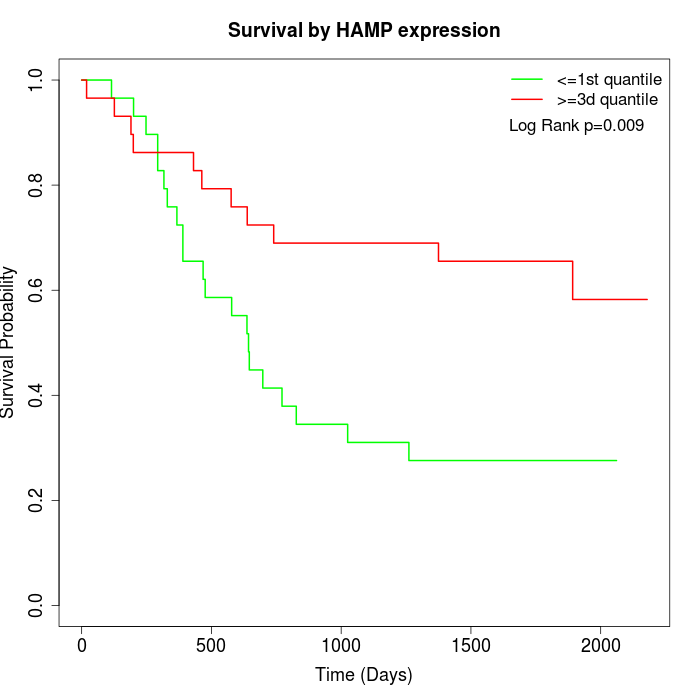
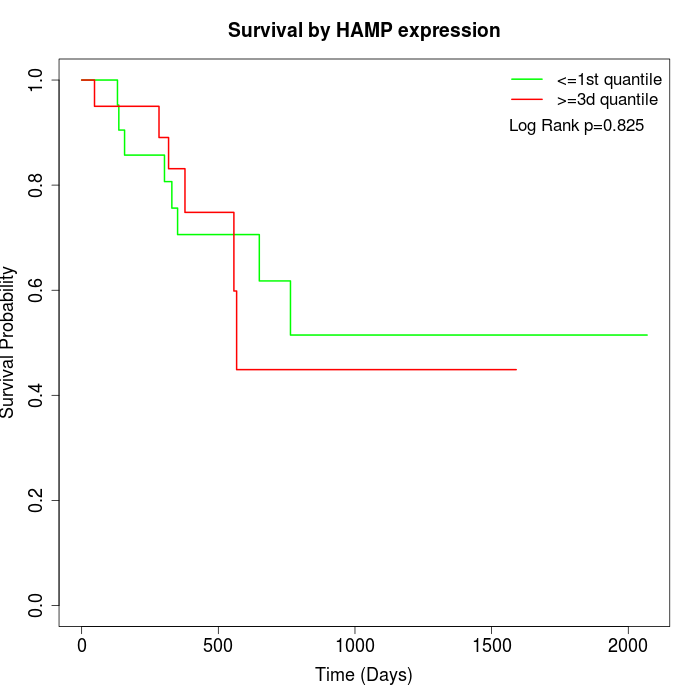
Survival by HAMP expression:
Note: Click image to view full size file.
Copy number change of HAMP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HAMP | 57817 | 6 | 4 | 20 | |
| GSE20123 | HAMP | 57817 | 6 | 3 | 21 | |
| GSE43470 | HAMP | 57817 | 3 | 7 | 33 | |
| GSE46452 | HAMP | 57817 | 48 | 1 | 10 | |
| GSE47630 | HAMP | 57817 | 9 | 5 | 26 | |
| GSE54993 | HAMP | 57817 | 17 | 3 | 50 | |
| GSE54994 | HAMP | 57817 | 8 | 9 | 36 | |
| GSE60625 | HAMP | 57817 | 9 | 0 | 2 | |
| GSE74703 | HAMP | 57817 | 3 | 4 | 29 | |
| GSE74704 | HAMP | 57817 | 6 | 1 | 13 | |
| TCGA | HAMP | 57817 | 21 | 11 | 64 |
Total number of gains: 136; Total number of losses: 48; Total Number of normals: 304.
Somatic mutations of HAMP:
Generating mutation plots.
Highly correlated genes for HAMP:
Showing top 20/387 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HAMP | RNF222 | 0.864461 | 3 | 0 | 3 |
| HAMP | IVL | 0.836616 | 3 | 0 | 3 |
| HAMP | GPR62 | 0.835727 | 3 | 0 | 3 |
| HAMP | NOTO | 0.806016 | 3 | 0 | 3 |
| HAMP | FSHR | 0.804002 | 3 | 0 | 3 |
| HAMP | KRTAP5-4 | 0.781588 | 3 | 0 | 3 |
| HAMP | CLEC18C | 0.738471 | 3 | 0 | 3 |
| HAMP | IGFL1 | 0.738047 | 3 | 0 | 3 |
| HAMP | DSC2 | 0.734194 | 3 | 0 | 3 |
| HAMP | OXT | 0.727887 | 4 | 0 | 4 |
| HAMP | KPRP | 0.725075 | 3 | 0 | 3 |
| HAMP | PHLDB3 | 0.7143 | 3 | 0 | 3 |
| HAMP | TMEM45B | 0.713773 | 3 | 0 | 3 |
| HAMP | C3orf20 | 0.712972 | 3 | 0 | 3 |
| HAMP | MFSD2B | 0.711219 | 3 | 0 | 3 |
| HAMP | HMGB4 | 0.709452 | 4 | 0 | 3 |
| HAMP | FAM163B | 0.706613 | 3 | 0 | 3 |
| HAMP | NCCRP1 | 0.705114 | 3 | 0 | 3 |
| HAMP | DNAI2 | 0.701417 | 4 | 0 | 3 |
| HAMP | DEGS2 | 0.701193 | 4 | 0 | 4 |
For details and further investigation, click here