| Full name: C-type lectin domain family 12 member B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.2 | ||
| Entrez ID: 387837 | HGNC ID: HGNC:31966 | Ensembl Gene: ENSG00000256660 | OMIM ID: 617573 |
| Drug and gene relationship at DGIdb | |||
Expression of CLEC12B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLEC12B | 387837 | 231357_at | -0.0197 | 0.9599 | |
| GSE26886 | CLEC12B | 387837 | 231357_at | 0.4283 | 0.0485 | |
| GSE45670 | CLEC12B | 387837 | 231357_at | 0.2223 | 0.0199 | |
| GSE53622 | CLEC12B | 387837 | 1479 | -0.7472 | 0.0000 | |
| GSE53624 | CLEC12B | 387837 | 1479 | -1.1022 | 0.0000 | |
| GSE63941 | CLEC12B | 387837 | 231357_at | -0.3992 | 0.0240 | |
| GSE77861 | CLEC12B | 387837 | 231600_at | -0.0934 | 0.4381 | |
| GSE97050 | CLEC12B | 387837 | A_33_P3410659 | -0.0416 | 0.8663 | |
| TCGA | CLEC12B | 387837 | RNAseq | 2.5873 | 0.0003 |
Upregulated datasets: 1; Downregulated datasets: 1.
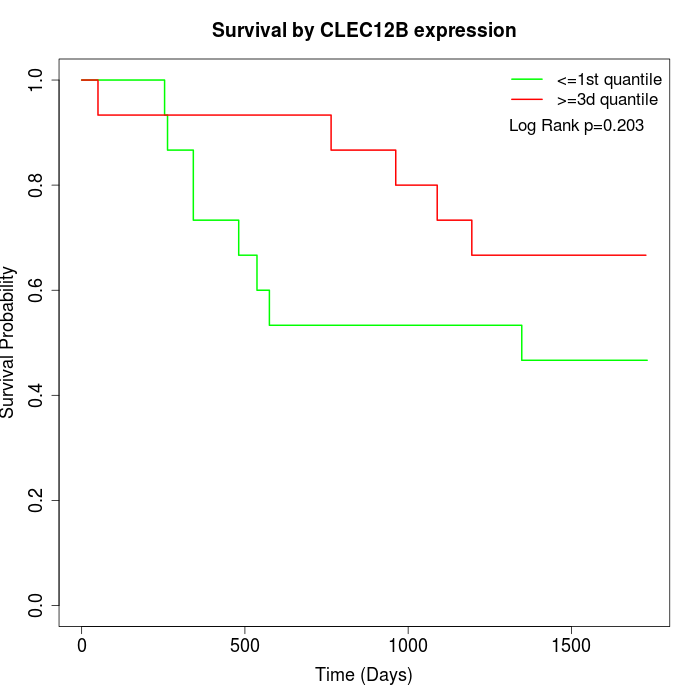
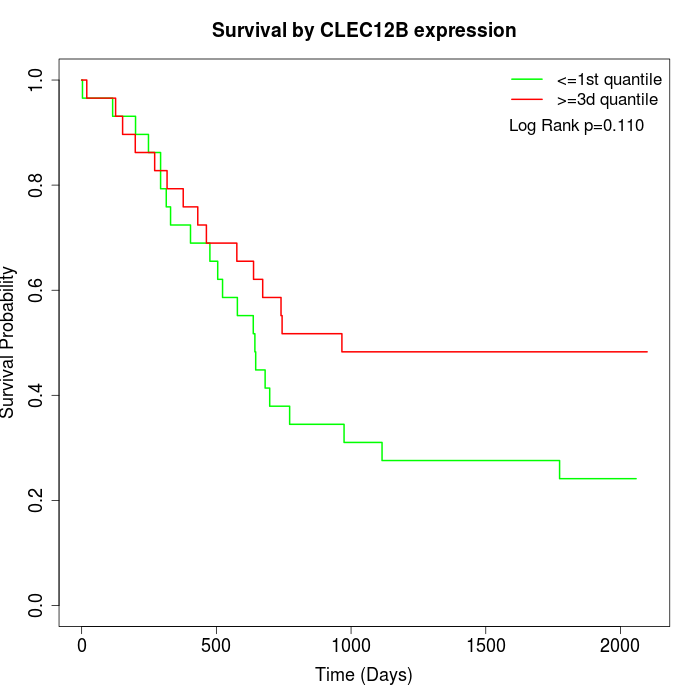
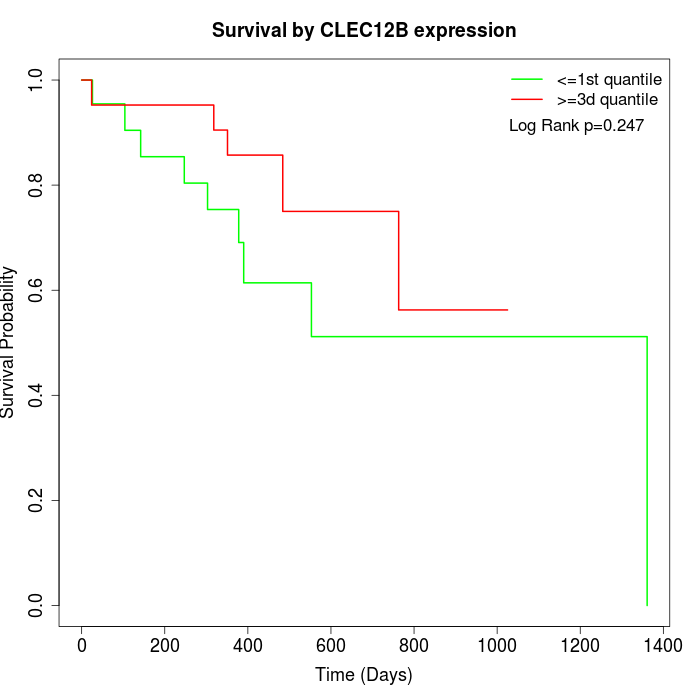
Survival by CLEC12B expression:
Note: Click image to view full size file.
Copy number change of CLEC12B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLEC12B | 387837 | 5 | 3 | 22 | |
| GSE20123 | CLEC12B | 387837 | 5 | 3 | 22 | |
| GSE43470 | CLEC12B | 387837 | 8 | 3 | 32 | |
| GSE46452 | CLEC12B | 387837 | 10 | 1 | 48 | |
| GSE47630 | CLEC12B | 387837 | 14 | 2 | 24 | |
| GSE54993 | CLEC12B | 387837 | 1 | 10 | 59 | |
| GSE54994 | CLEC12B | 387837 | 9 | 2 | 42 | |
| GSE60625 | CLEC12B | 387837 | 0 | 1 | 10 | |
| GSE74703 | CLEC12B | 387837 | 8 | 2 | 26 | |
| GSE74704 | CLEC12B | 387837 | 3 | 2 | 15 | |
| TCGA | CLEC12B | 387837 | 40 | 6 | 50 |
Total number of gains: 103; Total number of losses: 35; Total Number of normals: 350.
Somatic mutations of CLEC12B:
Generating mutation plots.
Highly correlated genes for CLEC12B:
Showing top 20/81 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLEC12B | DCD | 0.80634 | 3 | 0 | 3 |
| CLEC12B | NHLRC4 | 0.805956 | 4 | 0 | 3 |
| CLEC12B | MID1 | 0.781703 | 3 | 0 | 3 |
| CLEC12B | RNF222 | 0.781263 | 3 | 0 | 3 |
| CLEC12B | ZNF684 | 0.77735 | 3 | 0 | 3 |
| CLEC12B | GPR62 | 0.765043 | 4 | 0 | 3 |
| CLEC12B | SLC27A5 | 0.755663 | 3 | 0 | 3 |
| CLEC12B | BCDIN3D | 0.75504 | 3 | 0 | 3 |
| CLEC12B | NOTO | 0.748114 | 3 | 0 | 3 |
| CLEC12B | CLEC18C | 0.747179 | 3 | 0 | 3 |
| CLEC12B | MAGEL2 | 0.737512 | 3 | 0 | 3 |
| CLEC12B | AIRE | 0.726965 | 3 | 0 | 3 |
| CLEC12B | SLC10A6 | 0.724877 | 3 | 0 | 3 |
| CLEC12B | OSGIN1 | 0.719461 | 3 | 0 | 3 |
| CLEC12B | PAX8 | 0.71414 | 3 | 0 | 3 |
| CLEC12B | FRMPD2 | 0.713443 | 3 | 0 | 3 |
| CLEC12B | ITGA2B | 0.705826 | 3 | 0 | 3 |
| CLEC12B | ATP7B | 0.702622 | 3 | 0 | 3 |
| CLEC12B | KRTAP5-4 | 0.702073 | 3 | 0 | 3 |
| CLEC12B | BARHL2 | 0.70151 | 3 | 0 | 3 |
For details and further investigation, click here