| Full name: INSM transcriptional repressor 1 | Alias Symbol: IA-1|IA1 | ||
| Type: protein-coding gene | Cytoband: 20p11.23 | ||
| Entrez ID: 3642 | HGNC ID: HGNC:6090 | Ensembl Gene: ENSG00000173404 | OMIM ID: 600010 |
| Drug and gene relationship at DGIdb | |||
Expression of INSM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | INSM1 | 3642 | 206502_s_at | 0.3101 | 0.2130 | |
| GSE20347 | INSM1 | 3642 | 206502_s_at | -0.0009 | 0.9900 | |
| GSE23400 | INSM1 | 3642 | 206502_s_at | 0.0705 | 0.4858 | |
| GSE26886 | INSM1 | 3642 | 206502_s_at | 0.3994 | 0.0557 | |
| GSE29001 | INSM1 | 3642 | 206502_s_at | 0.3932 | 0.4499 | |
| GSE38129 | INSM1 | 3642 | 206502_s_at | 0.0208 | 0.9537 | |
| GSE45670 | INSM1 | 3642 | 206502_s_at | 0.4137 | 0.6029 | |
| GSE53622 | INSM1 | 3642 | 32029 | 0.3376 | 0.0808 | |
| GSE53624 | INSM1 | 3642 | 32029 | 0.0945 | 0.6096 | |
| TCGA | INSM1 | 3642 | RNAseq | -2.0688 | 0.0010 |
Upregulated datasets: 0; Downregulated datasets: 1.
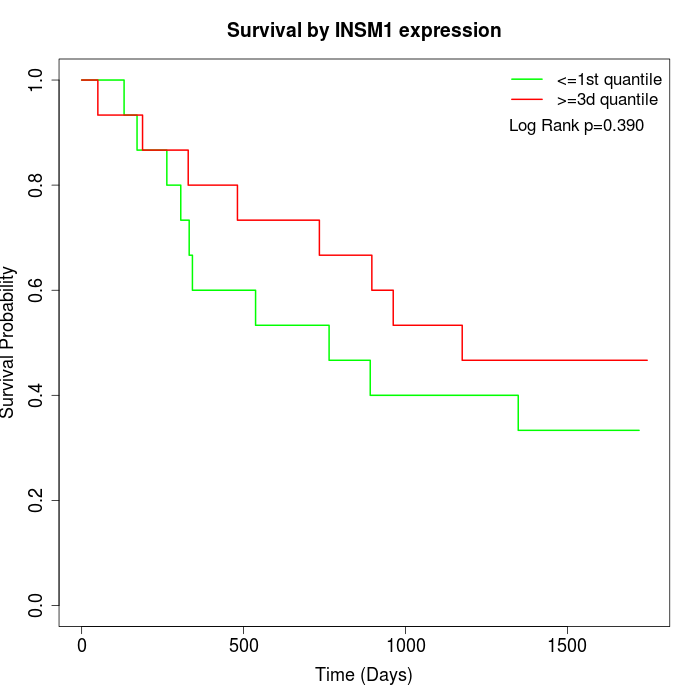
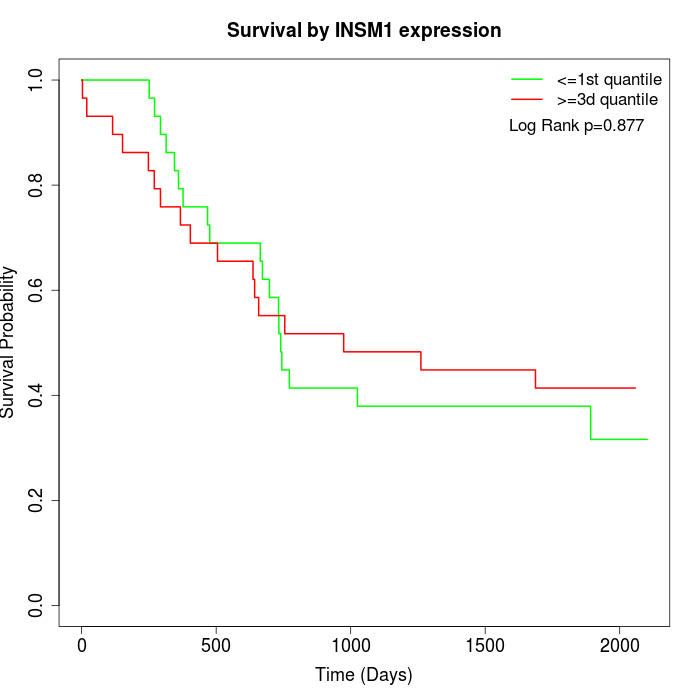
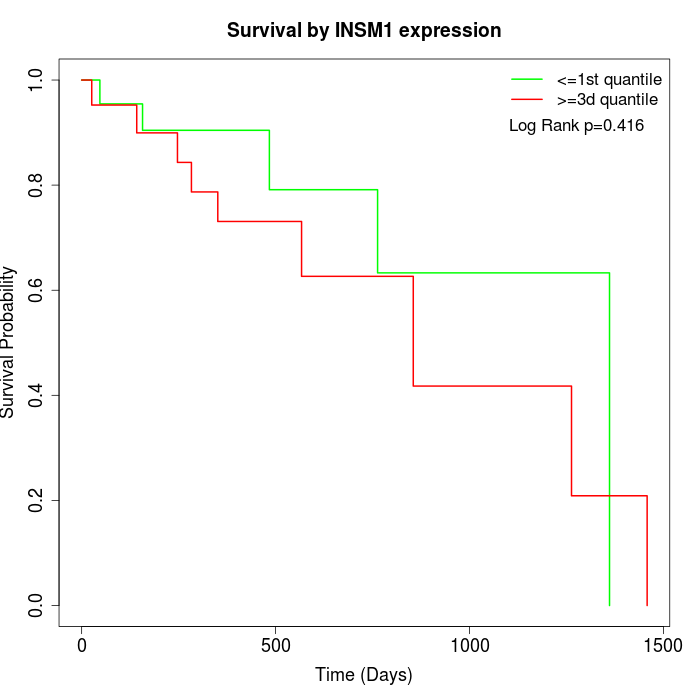
Survival by INSM1 expression:
Note: Click image to view full size file.
Copy number change of INSM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | INSM1 | 3642 | 13 | 2 | 15 | |
| GSE20123 | INSM1 | 3642 | 13 | 2 | 15 | |
| GSE43470 | INSM1 | 3642 | 11 | 1 | 31 | |
| GSE46452 | INSM1 | 3642 | 27 | 1 | 31 | |
| GSE47630 | INSM1 | 3642 | 17 | 4 | 19 | |
| GSE54993 | INSM1 | 3642 | 0 | 15 | 55 | |
| GSE54994 | INSM1 | 3642 | 27 | 2 | 24 | |
| GSE60625 | INSM1 | 3642 | 0 | 0 | 11 | |
| GSE74703 | INSM1 | 3642 | 8 | 1 | 27 | |
| GSE74704 | INSM1 | 3642 | 7 | 2 | 11 | |
| TCGA | INSM1 | 3642 | 43 | 11 | 42 |
Total number of gains: 166; Total number of losses: 41; Total Number of normals: 281.
Somatic mutations of INSM1:
Generating mutation plots.
Highly correlated genes for INSM1:
Showing top 20/47 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| INSM1 | TPH1 | 0.736435 | 3 | 0 | 3 |
| INSM1 | SLC5A6 | 0.723451 | 3 | 0 | 3 |
| INSM1 | IL13RA2 | 0.701312 | 4 | 0 | 3 |
| INSM1 | PAFAH1B3 | 0.696779 | 3 | 0 | 3 |
| INSM1 | RANBP17 | 0.695388 | 4 | 0 | 4 |
| INSM1 | TRO | 0.691509 | 3 | 0 | 3 |
| INSM1 | LRRTM4 | 0.690095 | 4 | 0 | 4 |
| INSM1 | GABRB1 | 0.680876 | 4 | 0 | 3 |
| INSM1 | ZNF180 | 0.673397 | 3 | 0 | 3 |
| INSM1 | RELB | 0.672603 | 5 | 0 | 4 |
| INSM1 | SH3GL2 | 0.672196 | 3 | 0 | 3 |
| INSM1 | RNF182 | 0.646286 | 3 | 0 | 3 |
| INSM1 | PTBP1 | 0.635607 | 3 | 0 | 3 |
| INSM1 | NGFR | 0.633952 | 3 | 0 | 3 |
| INSM1 | TUBB2B | 0.631533 | 5 | 0 | 4 |
| INSM1 | TYK2 | 0.630395 | 4 | 0 | 4 |
| INSM1 | IQCG | 0.622324 | 3 | 0 | 3 |
| INSM1 | HAUS7 | 0.620937 | 3 | 0 | 3 |
| INSM1 | KREMEN2 | 0.619083 | 3 | 0 | 3 |
| INSM1 | CLDN3 | 0.613334 | 4 | 0 | 3 |
For details and further investigation, click here