| Full name: leucine rich repeat transmembrane neuronal 4 | Alias Symbol: FLJ12568 | ||
| Type: protein-coding gene | Cytoband: 2p12 | ||
| Entrez ID: 80059 | HGNC ID: HGNC:19411 | Ensembl Gene: ENSG00000176204 | OMIM ID: 610870 |
| Drug and gene relationship at DGIdb | |||
Expression of LRRTM4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LRRTM4 | 80059 | 220345_at | 0.0031 | 0.9905 | |
| GSE20347 | LRRTM4 | 80059 | 220345_at | -0.0578 | 0.1910 | |
| GSE23400 | LRRTM4 | 80059 | 220345_at | -0.0242 | 0.1207 | |
| GSE26886 | LRRTM4 | 80059 | 220345_at | 0.1472 | 0.1015 | |
| GSE29001 | LRRTM4 | 80059 | 220345_at | -0.0232 | 0.8737 | |
| GSE38129 | LRRTM4 | 80059 | 220345_at | -0.0630 | 0.4803 | |
| GSE45670 | LRRTM4 | 80059 | 220345_at | 0.1017 | 0.4584 | |
| GSE53622 | LRRTM4 | 80059 | 7106 | -0.9379 | 0.0017 | |
| GSE53624 | LRRTM4 | 80059 | 7106 | -0.4893 | 0.0025 | |
| GSE63941 | LRRTM4 | 80059 | 220345_at | 0.0941 | 0.7336 | |
| GSE77861 | LRRTM4 | 80059 | 220345_at | -0.0187 | 0.8209 | |
| GSE97050 | LRRTM4 | 80059 | A_24_P60268 | -0.8973 | 0.1702 | |
| TCGA | LRRTM4 | 80059 | RNAseq | -0.7025 | 0.2539 |
Upregulated datasets: 0; Downregulated datasets: 0.
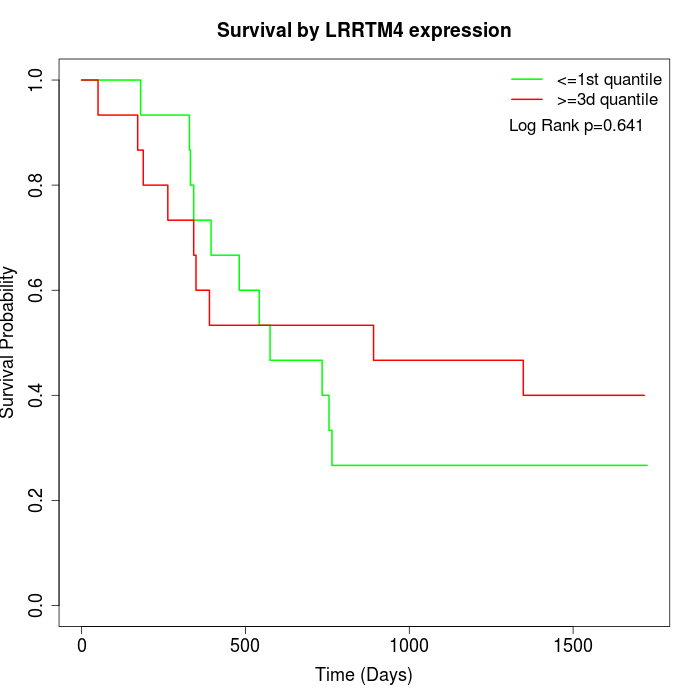
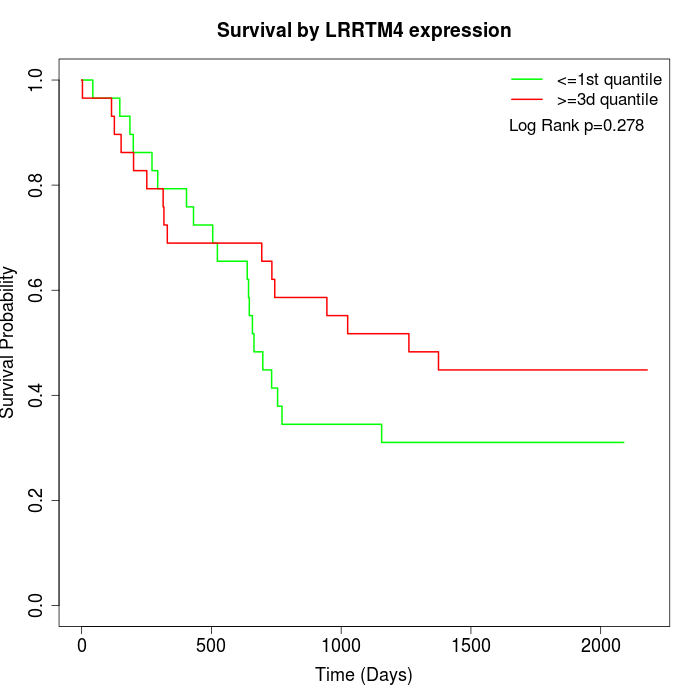
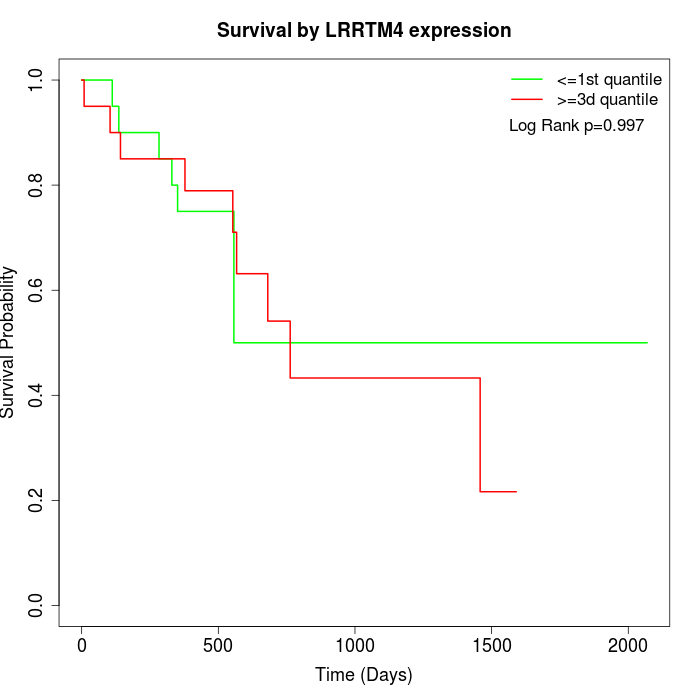
Survival by LRRTM4 expression:
Note: Click image to view full size file.
Copy number change of LRRTM4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LRRTM4 | 80059 | 11 | 3 | 16 | |
| GSE20123 | LRRTM4 | 80059 | 10 | 3 | 17 | |
| GSE43470 | LRRTM4 | 80059 | 4 | 0 | 39 | |
| GSE46452 | LRRTM4 | 80059 | 2 | 3 | 54 | |
| GSE47630 | LRRTM4 | 80059 | 7 | 0 | 33 | |
| GSE54993 | LRRTM4 | 80059 | 0 | 6 | 64 | |
| GSE54994 | LRRTM4 | 80059 | 10 | 0 | 43 | |
| GSE60625 | LRRTM4 | 80059 | 0 | 3 | 8 | |
| GSE74703 | LRRTM4 | 80059 | 4 | 0 | 32 | |
| GSE74704 | LRRTM4 | 80059 | 7 | 1 | 12 | |
| TCGA | LRRTM4 | 80059 | 35 | 4 | 57 |
Total number of gains: 90; Total number of losses: 23; Total Number of normals: 375.
Somatic mutations of LRRTM4:
Generating mutation plots.
Highly correlated genes for LRRTM4:
Showing top 20/65 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LRRTM4 | INSM1 | 0.690095 | 4 | 0 | 4 |
| LRRTM4 | FRMPD4 | 0.666202 | 3 | 0 | 3 |
| LRRTM4 | SPICE1 | 0.625757 | 3 | 0 | 3 |
| LRRTM4 | ZNF573 | 0.619767 | 3 | 0 | 3 |
| LRRTM4 | ZNF157 | 0.616931 | 3 | 0 | 3 |
| LRRTM4 | SH3GL2 | 0.60332 | 7 | 0 | 6 |
| LRRTM4 | STMN2 | 0.599601 | 5 | 0 | 4 |
| LRRTM4 | GLRB | 0.590433 | 4 | 0 | 3 |
| LRRTM4 | ARHGEF40 | 0.589328 | 3 | 0 | 3 |
| LRRTM4 | FABP7 | 0.589026 | 5 | 0 | 3 |
| LRRTM4 | DDX17 | 0.5888 | 3 | 0 | 3 |
| LRRTM4 | GRIK2 | 0.583862 | 6 | 0 | 5 |
| LRRTM4 | ENOX1 | 0.579216 | 6 | 0 | 4 |
| LRRTM4 | FAM193B | 0.576082 | 3 | 0 | 3 |
| LRRTM4 | CEP57L1 | 0.571911 | 4 | 0 | 3 |
| LRRTM4 | GABRB1 | 0.57083 | 7 | 0 | 4 |
| LRRTM4 | SHC2 | 0.570732 | 5 | 0 | 3 |
| LRRTM4 | YPEL1 | 0.563908 | 7 | 0 | 4 |
| LRRTM4 | ADCY2 | 0.563449 | 4 | 0 | 3 |
| LRRTM4 | AKAP12 | 0.56094 | 4 | 0 | 3 |
For details and further investigation, click here