| Full name: influenza virus NS1A binding protein | Alias Symbol: NS1-BP|HSPC068|NS-1|KIAA0850|ND1|KLHL39|ARA3 | ||
| Type: protein-coding gene | Cytoband: 1q25.3 | ||
| Entrez ID: 10625 | HGNC ID: HGNC:16951 | Ensembl Gene: ENSG00000116679 | OMIM ID: 609209 |
| Drug and gene relationship at DGIdb | |||
Expression of IVNS1ABP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IVNS1ABP | 10625 | 206245_s_at | 0.5342 | 0.1039 | |
| GSE20347 | IVNS1ABP | 10625 | 206245_s_at | 0.5582 | 0.0216 | |
| GSE23400 | IVNS1ABP | 10625 | 206245_s_at | 0.9057 | 0.0000 | |
| GSE26886 | IVNS1ABP | 10625 | 206245_s_at | 0.4765 | 0.0639 | |
| GSE29001 | IVNS1ABP | 10625 | 206245_s_at | 0.5757 | 0.0983 | |
| GSE38129 | IVNS1ABP | 10625 | 206245_s_at | 0.8808 | 0.0000 | |
| GSE45670 | IVNS1ABP | 10625 | 206245_s_at | 0.6127 | 0.0029 | |
| GSE53622 | IVNS1ABP | 10625 | 124466 | 0.2438 | 0.0038 | |
| GSE53624 | IVNS1ABP | 10625 | 124466 | 0.2552 | 0.0004 | |
| GSE63941 | IVNS1ABP | 10625 | 206245_s_at | 0.6137 | 0.4833 | |
| GSE77861 | IVNS1ABP | 10625 | 206245_s_at | 0.7840 | 0.0432 | |
| GSE97050 | IVNS1ABP | 10625 | A_23_P391506 | 0.3794 | 0.1894 | |
| SRP007169 | IVNS1ABP | 10625 | RNAseq | 0.6503 | 0.0971 | |
| SRP008496 | IVNS1ABP | 10625 | RNAseq | 1.1113 | 0.0001 | |
| SRP064894 | IVNS1ABP | 10625 | RNAseq | 0.9354 | 0.0007 | |
| SRP133303 | IVNS1ABP | 10625 | RNAseq | 0.7060 | 0.0003 | |
| SRP159526 | IVNS1ABP | 10625 | RNAseq | 0.4381 | 0.2050 | |
| SRP193095 | IVNS1ABP | 10625 | RNAseq | 0.3334 | 0.0072 | |
| SRP219564 | IVNS1ABP | 10625 | RNAseq | 0.7196 | 0.1482 | |
| TCGA | IVNS1ABP | 10625 | RNAseq | 0.0150 | 0.7725 |
Upregulated datasets: 1; Downregulated datasets: 0.
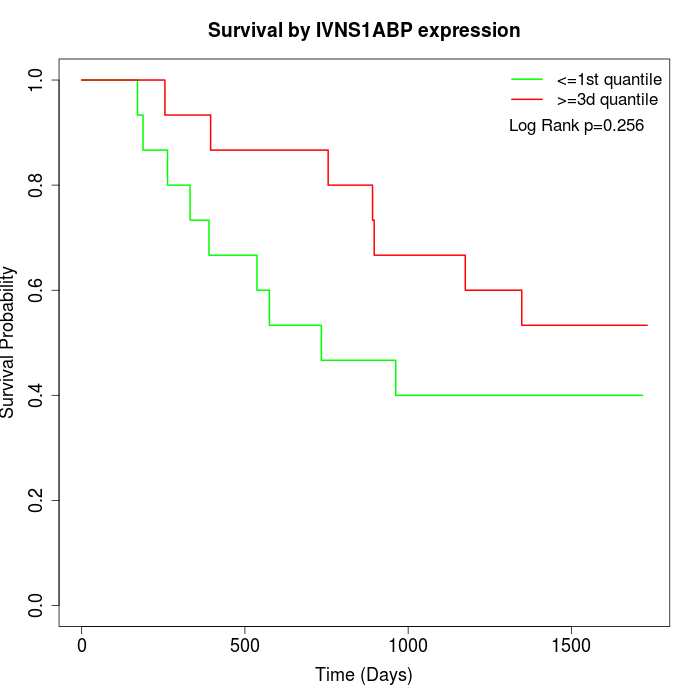
Survival by IVNS1ABP expression:
Note: Click image to view full size file.
Copy number change of IVNS1ABP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IVNS1ABP | 10625 | 12 | 0 | 18 | |
| GSE20123 | IVNS1ABP | 10625 | 12 | 0 | 18 | |
| GSE43470 | IVNS1ABP | 10625 | 7 | 1 | 35 | |
| GSE46452 | IVNS1ABP | 10625 | 3 | 1 | 55 | |
| GSE47630 | IVNS1ABP | 10625 | 14 | 0 | 26 | |
| GSE54993 | IVNS1ABP | 10625 | 0 | 6 | 64 | |
| GSE54994 | IVNS1ABP | 10625 | 15 | 0 | 38 | |
| GSE60625 | IVNS1ABP | 10625 | 0 | 0 | 11 | |
| GSE74703 | IVNS1ABP | 10625 | 7 | 1 | 28 | |
| GSE74704 | IVNS1ABP | 10625 | 5 | 0 | 15 | |
| TCGA | IVNS1ABP | 10625 | 40 | 4 | 52 |
Total number of gains: 115; Total number of losses: 13; Total Number of normals: 360.
Somatic mutations of IVNS1ABP:
Generating mutation plots.
Highly correlated genes for IVNS1ABP:
Showing top 20/983 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IVNS1ABP | NUDCD1 | 0.756589 | 4 | 0 | 4 |
| IVNS1ABP | PRKAB2 | 0.719783 | 3 | 0 | 3 |
| IVNS1ABP | MSANTD3 | 0.713989 | 3 | 0 | 3 |
| IVNS1ABP | FAM91A1 | 0.695386 | 4 | 0 | 4 |
| IVNS1ABP | CDCA2 | 0.694304 | 4 | 0 | 4 |
| IVNS1ABP | SALL4 | 0.679817 | 3 | 0 | 3 |
| IVNS1ABP | TRAF3 | 0.674252 | 6 | 0 | 5 |
| IVNS1ABP | DCUN1D5 | 0.669226 | 4 | 0 | 3 |
| IVNS1ABP | TMEM138 | 0.667897 | 5 | 0 | 4 |
| IVNS1ABP | C6orf141 | 0.667702 | 4 | 0 | 4 |
| IVNS1ABP | FOXK1 | 0.664295 | 4 | 0 | 3 |
| IVNS1ABP | WDR66 | 0.662657 | 5 | 0 | 4 |
| IVNS1ABP | HLA-A | 0.660268 | 5 | 0 | 5 |
| IVNS1ABP | HOXA10 | 0.659981 | 5 | 0 | 5 |
| IVNS1ABP | TDRD5 | 0.657461 | 4 | 0 | 4 |
| IVNS1ABP | DDX55 | 0.657319 | 5 | 0 | 4 |
| IVNS1ABP | LAMB3 | 0.657239 | 10 | 0 | 9 |
| IVNS1ABP | TYW5 | 0.656926 | 3 | 0 | 3 |
| IVNS1ABP | ZNF114 | 0.654405 | 3 | 0 | 3 |
| IVNS1ABP | CD276 | 0.651507 | 4 | 0 | 3 |
For details and further investigation, click here