| Full name: potassium channel tetramerization domain containing 20 | Alias Symbol: dJ108K11.3|MGC14254 | ||
| Type: protein-coding gene | Cytoband: 6p21.31 | ||
| Entrez ID: 222658 | HGNC ID: HGNC:21052 | Ensembl Gene: ENSG00000112078 | OMIM ID: 615932 |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD20:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD20 | 222658 | 228299_at | 0.2023 | 0.5194 | |
| GSE20347 | KCTD20 | 222658 | 214849_at | 0.0001 | 0.9995 | |
| GSE23400 | KCTD20 | 222658 | 214849_at | -0.2220 | 0.0000 | |
| GSE26886 | KCTD20 | 222658 | 228299_at | -0.3209 | 0.0981 | |
| GSE29001 | KCTD20 | 222658 | 214849_at | -0.3548 | 0.0144 | |
| GSE38129 | KCTD20 | 222658 | 214849_at | 0.0121 | 0.9035 | |
| GSE45670 | KCTD20 | 222658 | 228299_at | -0.3689 | 0.0060 | |
| GSE53622 | KCTD20 | 222658 | 85346 | -0.5075 | 0.0000 | |
| GSE53624 | KCTD20 | 222658 | 85346 | -0.7279 | 0.0000 | |
| GSE63941 | KCTD20 | 222658 | 228299_at | -1.0560 | 0.0197 | |
| GSE77861 | KCTD20 | 222658 | 228299_at | -0.6046 | 0.0847 | |
| GSE97050 | KCTD20 | 222658 | A_23_P374351 | -0.2552 | 0.3973 | |
| SRP007169 | KCTD20 | 222658 | RNAseq | -1.0846 | 0.0035 | |
| SRP008496 | KCTD20 | 222658 | RNAseq | -0.7288 | 0.0020 | |
| SRP064894 | KCTD20 | 222658 | RNAseq | -0.7888 | 0.0001 | |
| SRP133303 | KCTD20 | 222658 | RNAseq | -0.3991 | 0.0006 | |
| SRP159526 | KCTD20 | 222658 | RNAseq | -0.5141 | 0.0873 | |
| SRP193095 | KCTD20 | 222658 | RNAseq | -0.5835 | 0.0000 | |
| SRP219564 | KCTD20 | 222658 | RNAseq | -0.4359 | 0.1364 | |
| TCGA | KCTD20 | 222658 | RNAseq | -0.0602 | 0.3071 |
Upregulated datasets: 0; Downregulated datasets: 2.
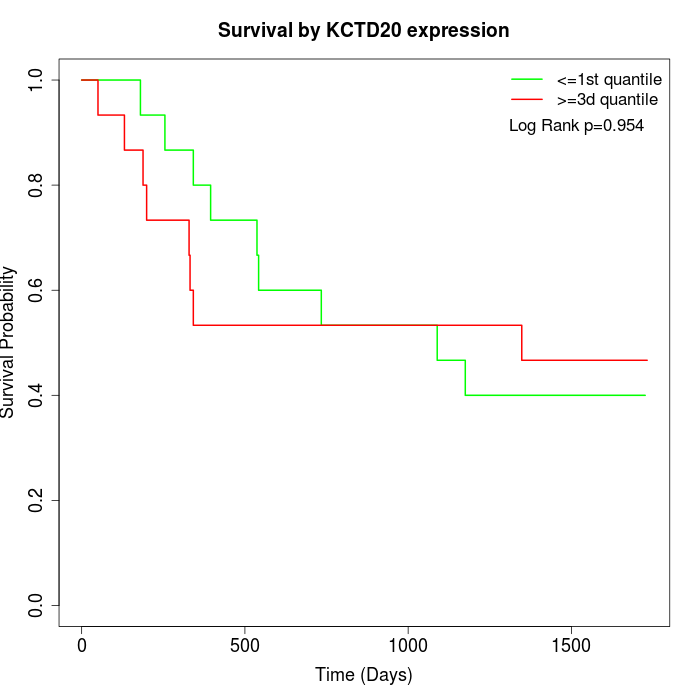
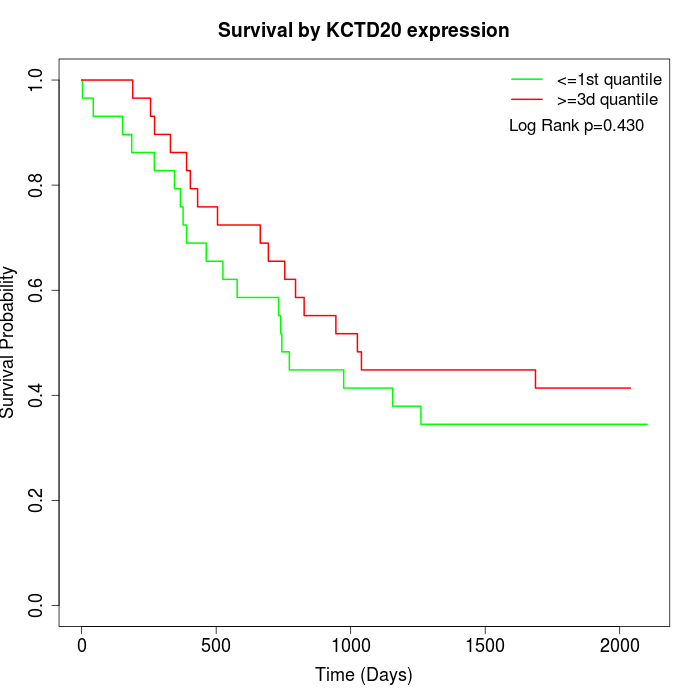
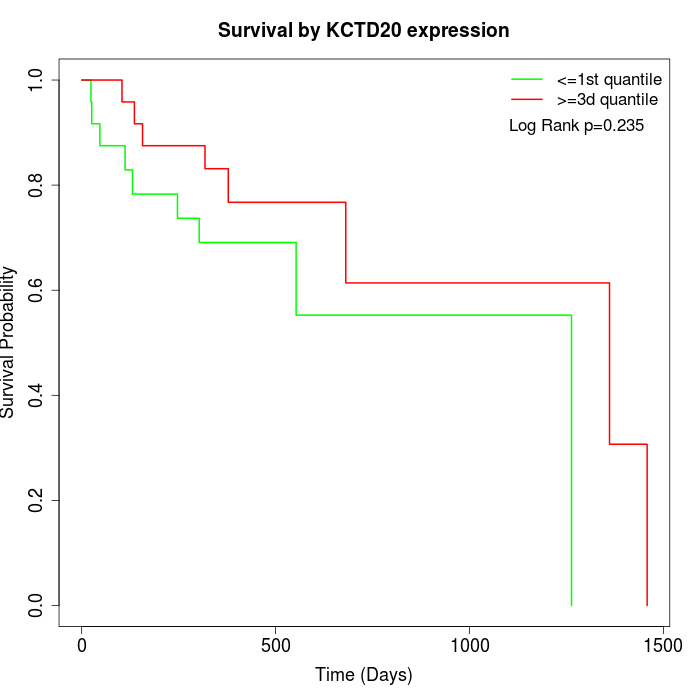
Survival by KCTD20 expression:
Note: Click image to view full size file.
Copy number change of KCTD20:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD20 | 222658 | 6 | 0 | 24 | |
| GSE20123 | KCTD20 | 222658 | 6 | 0 | 24 | |
| GSE43470 | KCTD20 | 222658 | 5 | 1 | 37 | |
| GSE46452 | KCTD20 | 222658 | 2 | 9 | 48 | |
| GSE47630 | KCTD20 | 222658 | 8 | 4 | 28 | |
| GSE54993 | KCTD20 | 222658 | 3 | 1 | 66 | |
| GSE54994 | KCTD20 | 222658 | 10 | 5 | 38 | |
| GSE60625 | KCTD20 | 222658 | 1 | 0 | 10 | |
| GSE74703 | KCTD20 | 222658 | 5 | 0 | 31 | |
| GSE74704 | KCTD20 | 222658 | 3 | 0 | 17 | |
| TCGA | KCTD20 | 222658 | 15 | 15 | 66 |
Total number of gains: 64; Total number of losses: 35; Total Number of normals: 389.
Somatic mutations of KCTD20:
Generating mutation plots.
Highly correlated genes for KCTD20:
Showing top 20/417 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD20 | MYZAP | 0.668509 | 3 | 0 | 3 |
| KCTD20 | AZU1 | 0.663977 | 4 | 0 | 3 |
| KCTD20 | THRB | 0.646941 | 4 | 0 | 4 |
| KCTD20 | KBTBD8 | 0.638826 | 3 | 0 | 3 |
| KCTD20 | BAHD1 | 0.629925 | 3 | 0 | 3 |
| KCTD20 | IL10RB | 0.627431 | 3 | 0 | 3 |
| KCTD20 | SCAMP2 | 0.625416 | 5 | 0 | 4 |
| KCTD20 | POP4 | 0.623228 | 3 | 0 | 3 |
| KCTD20 | ZNF382 | 0.623071 | 3 | 0 | 3 |
| KCTD20 | SH3BGRL2 | 0.621355 | 6 | 0 | 5 |
| KCTD20 | RNF208 | 0.619401 | 4 | 0 | 3 |
| KCTD20 | OXT | 0.619284 | 5 | 0 | 4 |
| KCTD20 | UBL3 | 0.614677 | 8 | 0 | 7 |
| KCTD20 | KRT78 | 0.613501 | 6 | 0 | 5 |
| KCTD20 | ANKRD13A | 0.609898 | 5 | 0 | 5 |
| KCTD20 | LYSMD3 | 0.609494 | 4 | 0 | 3 |
| KCTD20 | PBRM1 | 0.608599 | 6 | 0 | 5 |
| KCTD20 | ARSF | 0.606407 | 4 | 0 | 3 |
| KCTD20 | CAB39L | 0.606094 | 7 | 0 | 6 |
| KCTD20 | RASSF5 | 0.605794 | 6 | 0 | 5 |
For details and further investigation, click here