| Full name: calcium binding protein 39 like | Alias Symbol: bA103J18.3|FLJ12577|MO2L | ||
| Type: protein-coding gene | Cytoband: 13q14.2 | ||
| Entrez ID: 81617 | HGNC ID: HGNC:20290 | Ensembl Gene: ENSG00000102547 | OMIM ID: 612175 |
| Drug and gene relationship at DGIdb | |||
CAB39L involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa04152 | AMPK signaling pathway |
Expression of CAB39L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CAB39L | 81617 | 225914_s_at | -2.5465 | 0.0036 | |
| GSE20347 | CAB39L | 81617 | 221003_s_at | -0.6121 | 0.0000 | |
| GSE23400 | CAB39L | 81617 | 221003_s_at | -0.3763 | 0.0000 | |
| GSE26886 | CAB39L | 81617 | 225914_s_at | -2.3293 | 0.0000 | |
| GSE29001 | CAB39L | 81617 | 221003_s_at | -0.7145 | 0.0000 | |
| GSE38129 | CAB39L | 81617 | 221003_s_at | -0.6561 | 0.0000 | |
| GSE45670 | CAB39L | 81617 | 225914_s_at | -1.7510 | 0.0004 | |
| GSE53622 | CAB39L | 81617 | 44297 | -2.6088 | 0.0000 | |
| GSE53624 | CAB39L | 81617 | 44297 | -2.7300 | 0.0000 | |
| GSE63941 | CAB39L | 81617 | 221003_s_at | -0.6454 | 0.2773 | |
| GSE77861 | CAB39L | 81617 | 225914_s_at | -1.4549 | 0.0010 | |
| GSE97050 | CAB39L | 81617 | A_23_P204879 | -1.2628 | 0.0976 | |
| SRP007169 | CAB39L | 81617 | RNAseq | -4.4238 | 0.0000 | |
| SRP008496 | CAB39L | 81617 | RNAseq | -3.8163 | 0.0000 | |
| SRP064894 | CAB39L | 81617 | RNAseq | -2.0661 | 0.0000 | |
| SRP133303 | CAB39L | 81617 | RNAseq | -2.3351 | 0.0000 | |
| SRP159526 | CAB39L | 81617 | RNAseq | -2.6683 | 0.0000 | |
| SRP193095 | CAB39L | 81617 | RNAseq | -2.5791 | 0.0000 | |
| SRP219564 | CAB39L | 81617 | RNAseq | -1.4871 | 0.0543 | |
| TCGA | CAB39L | 81617 | RNAseq | -1.0267 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 13.
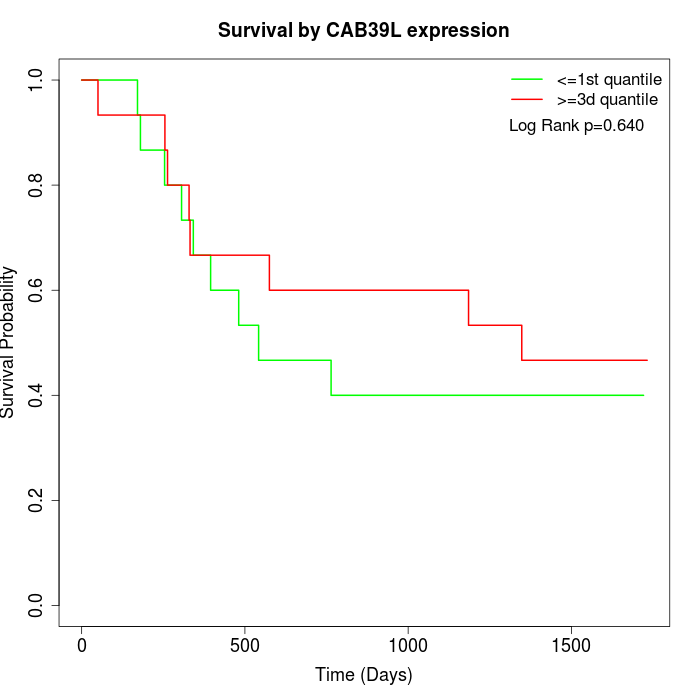
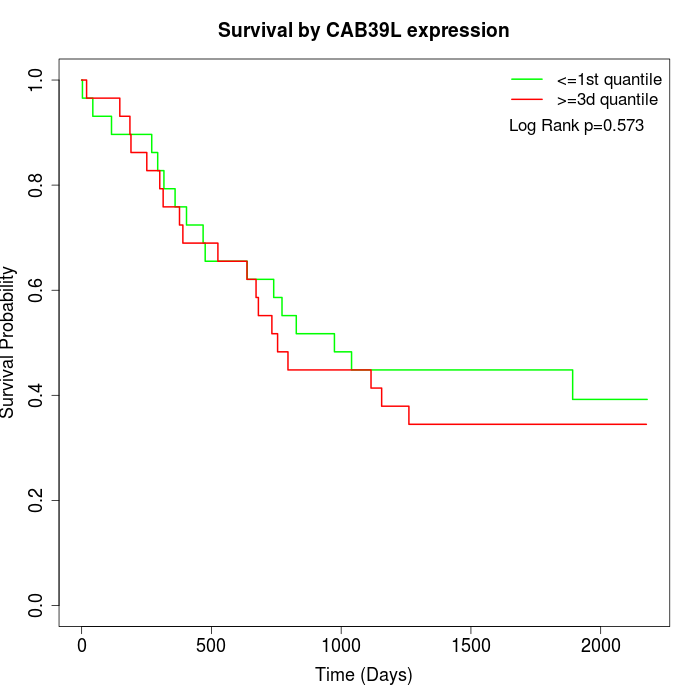
Survival by CAB39L expression:
Note: Click image to view full size file.
Copy number change of CAB39L:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CAB39L | 81617 | 1 | 13 | 16 | |
| GSE20123 | CAB39L | 81617 | 1 | 12 | 17 | |
| GSE43470 | CAB39L | 81617 | 3 | 13 | 27 | |
| GSE46452 | CAB39L | 81617 | 0 | 32 | 27 | |
| GSE47630 | CAB39L | 81617 | 2 | 27 | 11 | |
| GSE54993 | CAB39L | 81617 | 12 | 2 | 56 | |
| GSE54994 | CAB39L | 81617 | 2 | 14 | 37 | |
| GSE60625 | CAB39L | 81617 | 0 | 3 | 8 | |
| GSE74703 | CAB39L | 81617 | 3 | 10 | 23 | |
| GSE74704 | CAB39L | 81617 | 0 | 9 | 11 | |
| TCGA | CAB39L | 81617 | 9 | 37 | 50 |
Total number of gains: 33; Total number of losses: 172; Total Number of normals: 283.
Somatic mutations of CAB39L:
Generating mutation plots.
Highly correlated genes for CAB39L:
Showing top 20/2097 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CAB39L | SH3BGRL2 | 0.888238 | 7 | 0 | 7 |
| CAB39L | CGNL1 | 0.884907 | 7 | 0 | 7 |
| CAB39L | SNORA68 | 0.873017 | 4 | 0 | 4 |
| CAB39L | GBP6 | 0.86778 | 4 | 0 | 4 |
| CAB39L | SHROOM3 | 0.864972 | 7 | 0 | 7 |
| CAB39L | FCHO2 | 0.84851 | 7 | 0 | 7 |
| CAB39L | MPP7 | 0.846787 | 7 | 0 | 7 |
| CAB39L | SMIM5 | 0.844694 | 6 | 0 | 6 |
| CAB39L | FAM214A | 0.843878 | 7 | 0 | 7 |
| CAB39L | CLEC3B | 0.84384 | 3 | 0 | 3 |
| CAB39L | KAT2B | 0.839846 | 11 | 0 | 11 |
| CAB39L | GPD1L | 0.837793 | 11 | 0 | 11 |
| CAB39L | CYSRT1 | 0.831591 | 6 | 0 | 6 |
| CAB39L | ZNF662 | 0.831411 | 7 | 0 | 7 |
| CAB39L | EIF4E3 | 0.829272 | 7 | 0 | 7 |
| CAB39L | C5orf66-AS1 | 0.829089 | 6 | 0 | 6 |
| CAB39L | CYP21A2 | 0.825135 | 3 | 0 | 3 |
| CAB39L | HPGD | 0.821634 | 11 | 0 | 11 |
| CAB39L | SNX9 | 0.82046 | 7 | 0 | 7 |
| CAB39L | PAQR8 | 0.818921 | 7 | 0 | 7 |
For details and further investigation, click here