| Full name: Ras association domain family member 5 | Alias Symbol: Maxp1|NORE1|RAPL | ||
| Type: protein-coding gene | Cytoband: 1q32.1 | ||
| Entrez ID: 83593 | HGNC ID: HGNC:17609 | Ensembl Gene: ENSG00000266094 | OMIM ID: 607020 |
| Drug and gene relationship at DGIdb | |||
RASSF5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway | |
| hsa04015 | Rap1 signaling pathway | |
| hsa04670 | Leukocyte transendothelial migration | |
| hsa05200 | Pathways in cancer | |
| hsa05223 | Non-small cell lung cancer |
Expression of RASSF5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RASSF5 | 83593 | 223322_at | -0.4089 | 0.5512 | |
| GSE26886 | RASSF5 | 83593 | 223322_at | -1.7961 | 0.0000 | |
| GSE45670 | RASSF5 | 83593 | 223322_at | -0.6186 | 0.0291 | |
| GSE53622 | RASSF5 | 83593 | 31778 | -1.2858 | 0.0000 | |
| GSE53624 | RASSF5 | 83593 | 70501 | -1.7617 | 0.0000 | |
| GSE63941 | RASSF5 | 83593 | 223322_at | -0.0140 | 0.9902 | |
| GSE77861 | RASSF5 | 83593 | 223322_at | -0.6318 | 0.0013 | |
| GSE97050 | RASSF5 | 83593 | A_24_P336584 | -0.1860 | 0.8108 | |
| SRP007169 | RASSF5 | 83593 | RNAseq | -3.2275 | 0.0000 | |
| SRP008496 | RASSF5 | 83593 | RNAseq | -2.8637 | 0.0000 | |
| SRP064894 | RASSF5 | 83593 | RNAseq | -2.0456 | 0.0000 | |
| SRP133303 | RASSF5 | 83593 | RNAseq | -1.2341 | 0.0000 | |
| SRP159526 | RASSF5 | 83593 | RNAseq | -1.3660 | 0.0320 | |
| SRP193095 | RASSF5 | 83593 | RNAseq | -1.0519 | 0.0000 | |
| SRP219564 | RASSF5 | 83593 | RNAseq | -1.5775 | 0.0050 | |
| TCGA | RASSF5 | 83593 | RNAseq | 0.0650 | 0.5141 |
Upregulated datasets: 0; Downregulated datasets: 10.
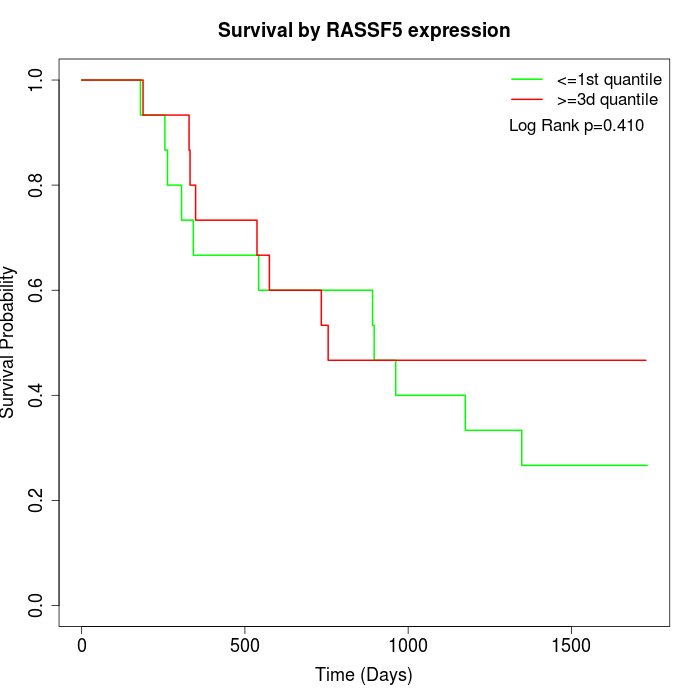
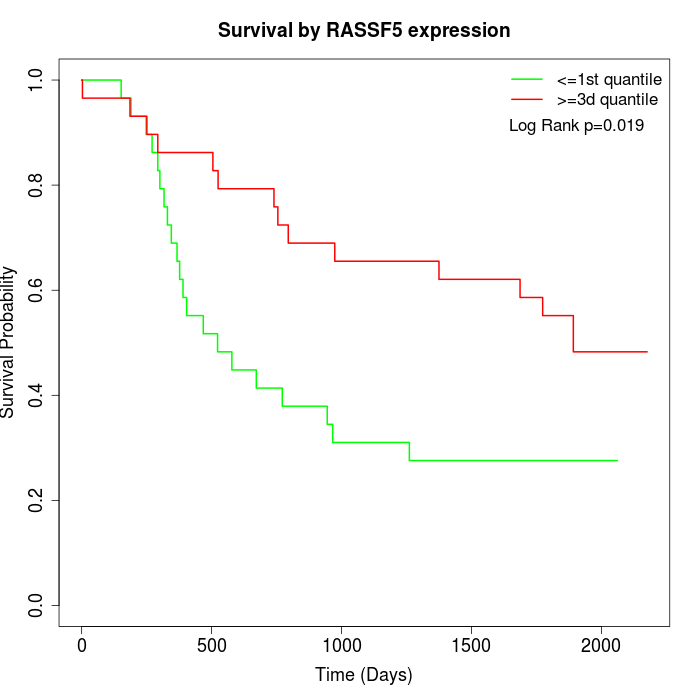
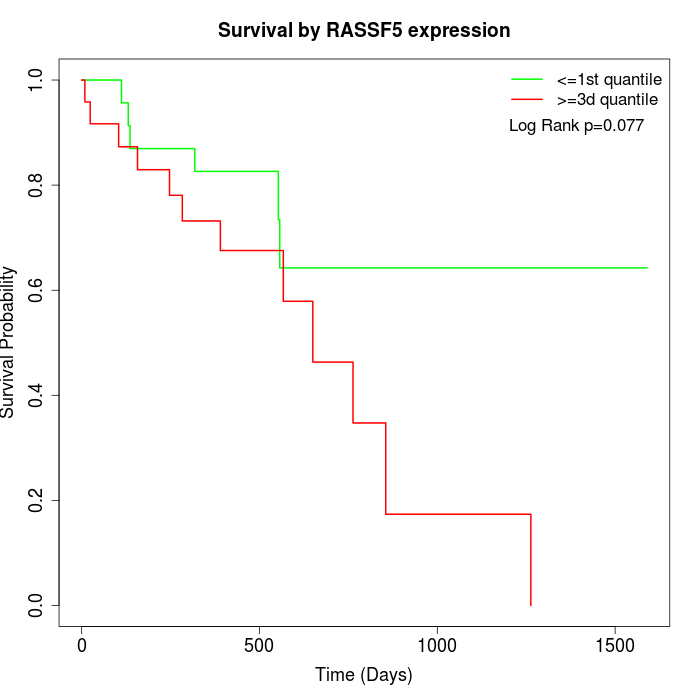
Survival by RASSF5 expression:
Note: Click image to view full size file.
Copy number change of RASSF5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RASSF5 | 83593 | 11 | 0 | 19 | |
| GSE20123 | RASSF5 | 83593 | 11 | 0 | 19 | |
| GSE43470 | RASSF5 | 83593 | 7 | 0 | 36 | |
| GSE46452 | RASSF5 | 83593 | 3 | 1 | 55 | |
| GSE47630 | RASSF5 | 83593 | 14 | 0 | 26 | |
| GSE54993 | RASSF5 | 83593 | 0 | 6 | 64 | |
| GSE54994 | RASSF5 | 83593 | 14 | 0 | 39 | |
| GSE60625 | RASSF5 | 83593 | 0 | 0 | 11 | |
| GSE74703 | RASSF5 | 83593 | 7 | 0 | 29 | |
| GSE74704 | RASSF5 | 83593 | 5 | 0 | 15 | |
| TCGA | RASSF5 | 83593 | 43 | 7 | 46 |
Total number of gains: 115; Total number of losses: 14; Total Number of normals: 359.
Somatic mutations of RASSF5:
Generating mutation plots.
Highly correlated genes for RASSF5:
Showing top 20/1778 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RASSF5 | ORMDL2 | 0.83563 | 4 | 0 | 4 |
| RASSF5 | RAB25 | 0.832899 | 7 | 0 | 7 |
| RASSF5 | VAV3 | 0.828113 | 5 | 0 | 5 |
| RASSF5 | RANBP9 | 0.827385 | 7 | 0 | 7 |
| RASSF5 | ARHGAP27 | 0.827178 | 7 | 0 | 7 |
| RASSF5 | USP6NL | 0.826671 | 7 | 0 | 7 |
| RASSF5 | RALGPS2 | 0.822484 | 7 | 0 | 7 |
| RASSF5 | TIAM1 | 0.820556 | 7 | 0 | 7 |
| RASSF5 | TMEM9B | 0.813014 | 7 | 0 | 7 |
| RASSF5 | CCDC6 | 0.809562 | 7 | 0 | 7 |
| RASSF5 | UBL3 | 0.809445 | 6 | 0 | 6 |
| RASSF5 | EPS8L2 | 0.809067 | 7 | 0 | 7 |
| RASSF5 | RBM47 | 0.801161 | 7 | 0 | 7 |
| RASSF5 | PHACTR4 | 0.800436 | 7 | 0 | 7 |
| RASSF5 | C5orf66-AS1 | 0.800264 | 6 | 0 | 6 |
| RASSF5 | EHD3 | 0.79977 | 7 | 0 | 7 |
| RASSF5 | PCBP1-AS1 | 0.797625 | 3 | 0 | 3 |
| RASSF5 | SLC16A6 | 0.795865 | 7 | 0 | 7 |
| RASSF5 | RNF141 | 0.794831 | 7 | 0 | 7 |
| RASSF5 | TP53I3 | 0.794389 | 7 | 0 | 7 |
For details and further investigation, click here