| Full name: L3MBTL histone methyl-lysine binding protein 1 | Alias Symbol: ZC2HC3|dJ138B7.3|DKFZp586P1522|KIAA0681 | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 26013 | HGNC ID: HGNC:15905 | Ensembl Gene: ENSG00000185513 | OMIM ID: 608802 |
| Drug and gene relationship at DGIdb | |||
Expression of L3MBTL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | L3MBTL1 | 26013 | 216076_at | 0.0994 | 0.6355 | |
| GSE20347 | L3MBTL1 | 26013 | 216076_at | -0.1049 | 0.2462 | |
| GSE23400 | L3MBTL1 | 26013 | 213837_at | -0.1473 | 0.0017 | |
| GSE26886 | L3MBTL1 | 26013 | 216076_at | 0.1783 | 0.1391 | |
| GSE29001 | L3MBTL1 | 26013 | 213837_at | -0.3419 | 0.0057 | |
| GSE38129 | L3MBTL1 | 26013 | 216076_at | -0.1682 | 0.1554 | |
| GSE45670 | L3MBTL1 | 26013 | 210306_at | -0.1287 | 0.7003 | |
| GSE53622 | L3MBTL1 | 26013 | 62082 | 0.2028 | 0.0598 | |
| GSE53624 | L3MBTL1 | 26013 | 119570 | -0.2072 | 0.0816 | |
| GSE63941 | L3MBTL1 | 26013 | 216076_at | 0.1917 | 0.1429 | |
| GSE77861 | L3MBTL1 | 26013 | 216076_at | -0.1288 | 0.4438 | |
| GSE97050 | L3MBTL1 | A_19_P00316107 | 0.0226 | 0.9347 | ||
| SRP007169 | L3MBTL1 | 26013 | RNAseq | 1.7140 | 0.0678 | |
| SRP064894 | L3MBTL1 | 26013 | RNAseq | -0.1536 | 0.6290 | |
| SRP133303 | L3MBTL1 | 26013 | RNAseq | -0.0735 | 0.7892 | |
| SRP159526 | L3MBTL1 | 26013 | RNAseq | -0.1640 | 0.6630 | |
| SRP193095 | L3MBTL1 | 26013 | RNAseq | 0.1740 | 0.2845 | |
| SRP219564 | L3MBTL1 | 26013 | RNAseq | 0.1032 | 0.8651 |
Upregulated datasets: 0; Downregulated datasets: 0.
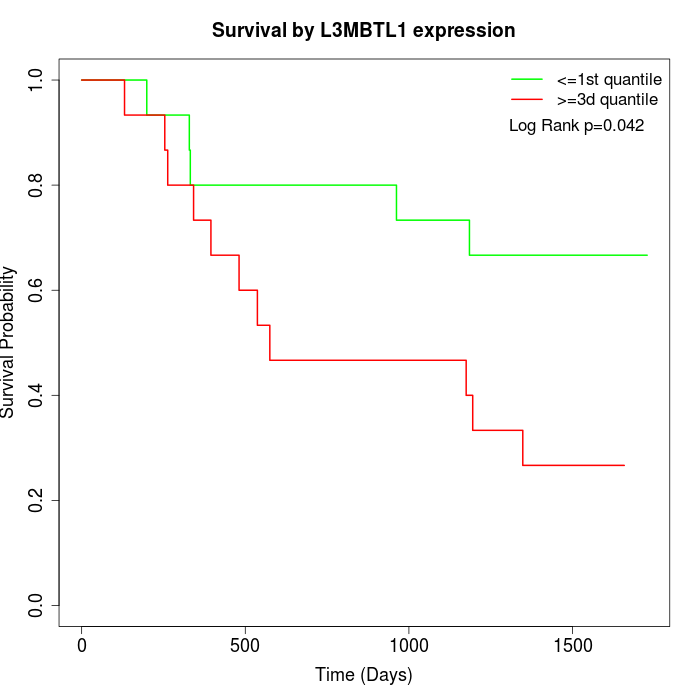
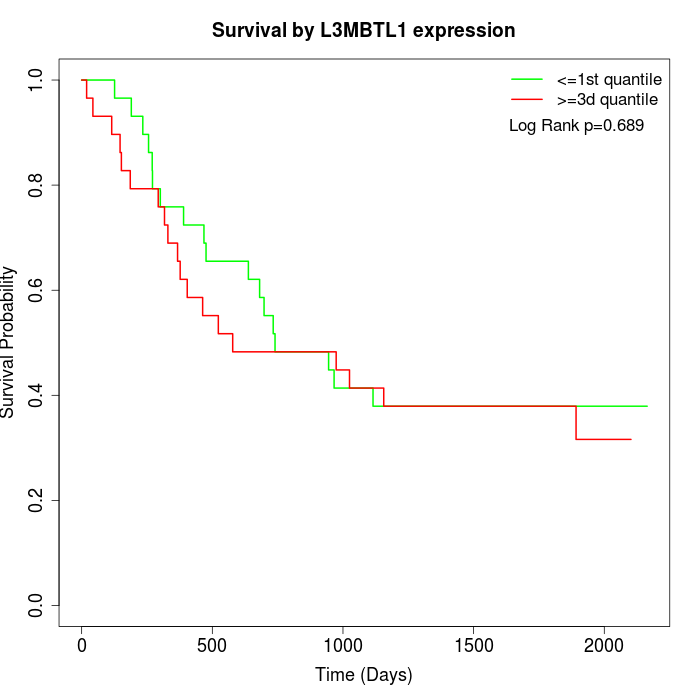
Survival by L3MBTL1 expression:
Note: Click image to view full size file.
Copy number change of L3MBTL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | L3MBTL1 | 26013 | 13 | 1 | 16 | |
| GSE20123 | L3MBTL1 | 26013 | 13 | 1 | 16 | |
| GSE43470 | L3MBTL1 | 26013 | 12 | 1 | 30 | |
| GSE46452 | L3MBTL1 | 26013 | 29 | 0 | 30 | |
| GSE47630 | L3MBTL1 | 26013 | 24 | 0 | 16 | |
| GSE54993 | L3MBTL1 | 26013 | 0 | 17 | 53 | |
| GSE54994 | L3MBTL1 | 26013 | 25 | 0 | 28 | |
| GSE60625 | L3MBTL1 | 26013 | 0 | 0 | 11 | |
| GSE74703 | L3MBTL1 | 26013 | 10 | 1 | 25 | |
| GSE74704 | L3MBTL1 | 26013 | 9 | 0 | 11 | |
| TCGA | L3MBTL1 | 26013 | 47 | 2 | 47 |
Total number of gains: 182; Total number of losses: 23; Total Number of normals: 283.
Somatic mutations of L3MBTL1:
Generating mutation plots.
Highly correlated genes for L3MBTL1:
Showing top 20/501 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| L3MBTL1 | ZC4H2 | 0.753467 | 3 | 0 | 3 |
| L3MBTL1 | BAZ2A | 0.746044 | 3 | 0 | 3 |
| L3MBTL1 | ARHGAP30 | 0.737563 | 4 | 0 | 4 |
| L3MBTL1 | NOXA1 | 0.735624 | 3 | 0 | 3 |
| L3MBTL1 | SCUBE1 | 0.72017 | 4 | 0 | 3 |
| L3MBTL1 | ADAMTS6 | 0.717342 | 4 | 0 | 3 |
| L3MBTL1 | KRTAP2-1 | 0.709823 | 3 | 0 | 3 |
| L3MBTL1 | ZDHHC19 | 0.708035 | 3 | 0 | 3 |
| L3MBTL1 | SLC22A31 | 0.70391 | 4 | 0 | 4 |
| L3MBTL1 | THPO | 0.702504 | 3 | 0 | 3 |
| L3MBTL1 | PNMA3 | 0.6971 | 7 | 0 | 6 |
| L3MBTL1 | NKX6-3 | 0.696269 | 3 | 0 | 3 |
| L3MBTL1 | USHBP1 | 0.689414 | 4 | 0 | 4 |
| L3MBTL1 | DIO3 | 0.686088 | 5 | 0 | 5 |
| L3MBTL1 | CLSTN3 | 0.682977 | 4 | 0 | 3 |
| L3MBTL1 | RCOR2 | 0.681216 | 4 | 0 | 3 |
| L3MBTL1 | RASL10B | 0.679014 | 5 | 0 | 4 |
| L3MBTL1 | FXYD2 | 0.678164 | 4 | 0 | 3 |
| L3MBTL1 | NPEPL1 | 0.677076 | 7 | 0 | 5 |
| L3MBTL1 | GPR78 | 0.672891 | 3 | 0 | 3 |
For details and further investigation, click here