| Full name: long intergenic non-protein coding RNA 886 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 3q25.31 | ||
| Entrez ID: 730091 | HGNC ID: HGNC:48572 | Ensembl Gene: ENSG00000240875 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00886:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00886 | 730091 | 230780_at | 0.0006 | 0.9992 | |
| GSE26886 | LINC00886 | 730091 | 230780_at | 0.3515 | 0.0399 | |
| GSE45670 | LINC00886 | 730091 | 230780_at | 0.0029 | 0.9917 | |
| GSE53622 | LINC00886 | 730091 | 90975 | -0.3761 | 0.0514 | |
| GSE53624 | LINC00886 | 730091 | 90975 | -0.2856 | 0.1804 | |
| GSE63941 | LINC00886 | 730091 | 230780_at | -0.2106 | 0.4127 | |
| GSE77861 | LINC00886 | 730091 | 230780_at | -0.1142 | 0.5956 | |
| SRP064894 | LINC00886 | 730091 | RNAseq | 0.3264 | 0.3337 | |
| SRP133303 | LINC00886 | 730091 | RNAseq | 0.0482 | 0.8709 | |
| SRP159526 | LINC00886 | 730091 | RNAseq | -0.9270 | 0.1067 | |
| SRP193095 | LINC00886 | 730091 | RNAseq | 0.2823 | 0.1286 | |
| SRP219564 | LINC00886 | 730091 | RNAseq | -0.5928 | 0.3867 |
Upregulated datasets: 0; Downregulated datasets: 0.
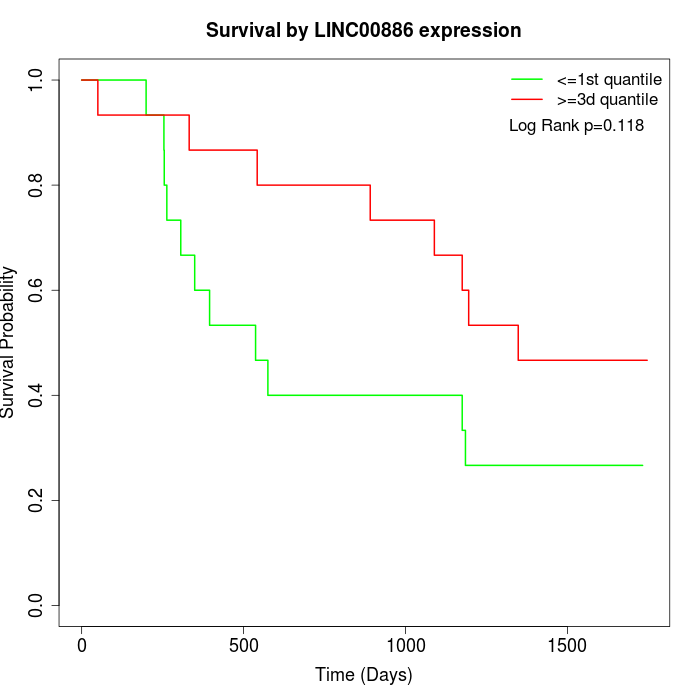
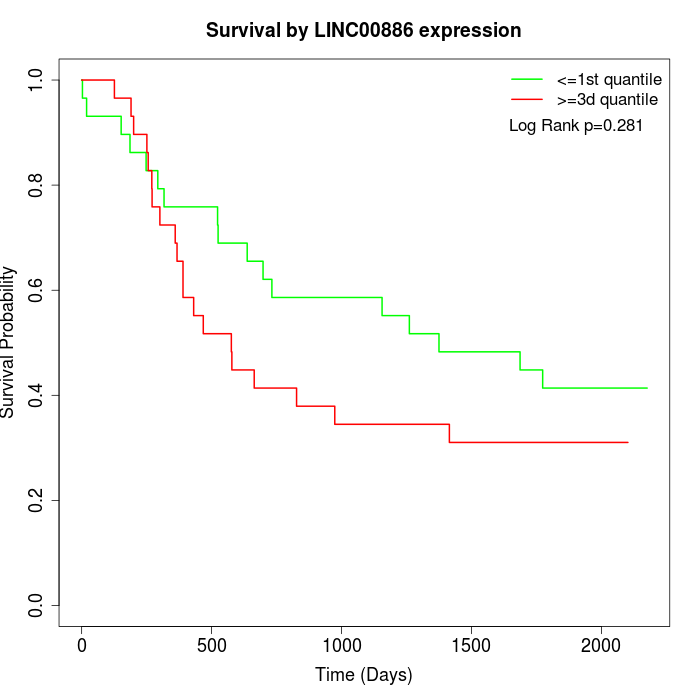
Survival by LINC00886 expression:
Note: Click image to view full size file.
Copy number change of LINC00886:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00886 | 730091 | 21 | 1 | 8 | |
| GSE20123 | LINC00886 | 730091 | 21 | 1 | 8 | |
| GSE43470 | LINC00886 | 730091 | 23 | 0 | 20 | |
| GSE46452 | LINC00886 | 730091 | 18 | 2 | 39 | |
| GSE47630 | LINC00886 | 730091 | 19 | 3 | 18 | |
| GSE54993 | LINC00886 | 730091 | 1 | 11 | 58 | |
| GSE54994 | LINC00886 | 730091 | 38 | 1 | 14 | |
| GSE60625 | LINC00886 | 730091 | 0 | 6 | 5 | |
| GSE74703 | LINC00886 | 730091 | 20 | 0 | 16 | |
| GSE74704 | LINC00886 | 730091 | 14 | 0 | 6 | |
| TCGA | LINC00886 | 730091 | 74 | 1 | 21 |
Total number of gains: 249; Total number of losses: 26; Total Number of normals: 213.
Somatic mutations of LINC00886:
Generating mutation plots.
Highly correlated genes for LINC00886:
Showing all 17 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00886 | ARVCF | 0.667757 | 3 | 0 | 3 |
| LINC00886 | CDC42EP2 | 0.65428 | 3 | 0 | 3 |
| LINC00886 | TTC9B | 0.653843 | 3 | 0 | 3 |
| LINC00886 | MIOX | 0.649945 | 3 | 0 | 3 |
| LINC00886 | RTEL1 | 0.636558 | 3 | 0 | 3 |
| LINC00886 | CLEC4M | 0.622256 | 3 | 0 | 3 |
| LINC00886 | TRIM10 | 0.611162 | 3 | 0 | 3 |
| LINC00886 | ATXN1L | 0.604441 | 3 | 0 | 3 |
| LINC00886 | PCBP4 | 0.59396 | 3 | 0 | 3 |
| LINC00886 | SLC22A25 | 0.588726 | 4 | 0 | 3 |
| LINC00886 | KCNIP3 | 0.582493 | 3 | 0 | 3 |
| LINC00886 | SHISA4 | 0.55779 | 3 | 0 | 3 |
| LINC00886 | PEAR1 | 0.528186 | 4 | 0 | 3 |
| LINC00886 | BEND4 | 0.519683 | 3 | 0 | 3 |
| LINC00886 | CLRN3 | 0.517002 | 4 | 0 | 3 |
| LINC00886 | CNIH2 | 0.512509 | 3 | 0 | 3 |
| LINC00886 | GIGYF2 | 0.507942 | 4 | 0 | 3 |
For details and further investigation, click here